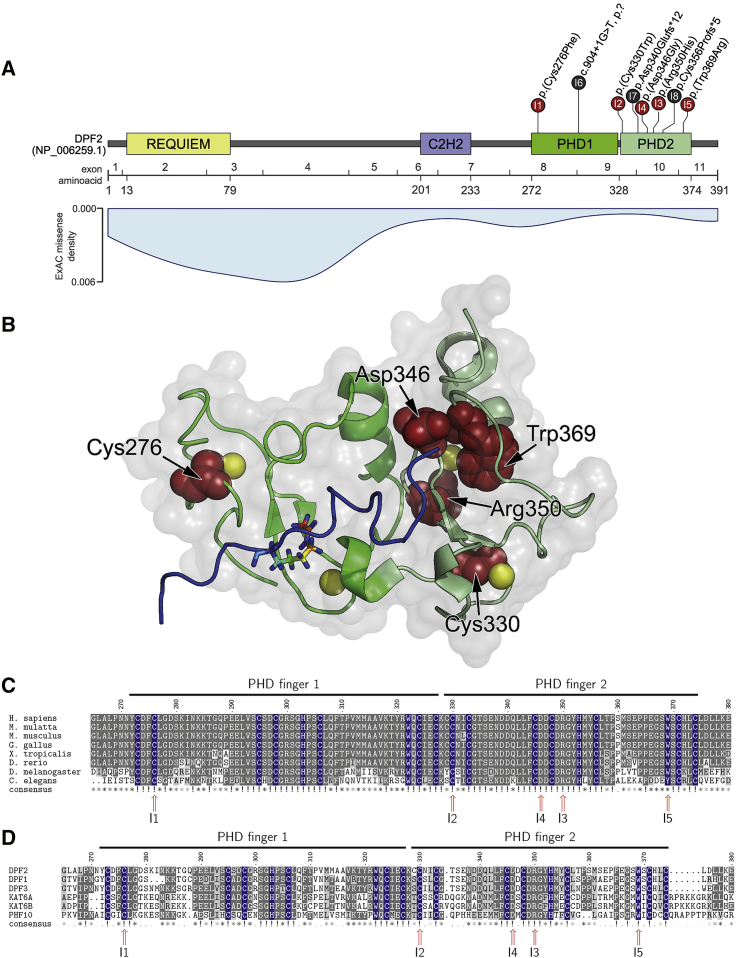
Figure 1.
Clustering of DPF2 Variants in PHD Fingers
(A) Schematic representation of DPF2, its domains (based on GenBank: NP_006259.1), the encoding exons (numbering based on GenBank: NM_006268.4), and the localization of DPF2 variants. Missense variants are presented in red, and truncating variants are in black. Note that the premature termination codons of the two truncating variants p.Asp340Glufs∗12 and p.Cys356Profs∗5 reside within 50 nucleotides upstream of the most 3′ exon-exon junction. The numbers in the circles indicate the affected individuals. For individual 6, c.904+1G>T is described at the genomic level because no RT-PCR could be performed. In light blue, a density blot of all missense variants reported in ExAC Browser version 0.3.1 shows markedly low frequency of variants in the PHD zinc fingers.
(B) The crystal structure of the DPF2 double PHD finger bound to a histone peptide containing acetylation at lysine 14 (H3K14ac) (PDB: 5B7918 and 2KWJ19) shows the clustering of the herein described missense variants in the tandem PHD finger domain (PHD finger 1 is colored in bright green, and PHD finger 2 is colored in pale green). The histone H3 backbone in the DPF2 binding pocket is shown in blue with an acetylated lysine residue at position 14 in stick representation. Zinc ions are represented as yellow spheres. The five affected amino acid residues are colored in red. The electrostatic surface is represented in gray with 80% opacity. Cys276 and Asp346 reside at the protein surface, whereas Cys330, Arg350, and Trp369 are buried in the PHD2 domain.20
(C) Multiple-sequence alignment of DPF2 orthologs at the de novo missense variant positions in the tandem PHD fingers shows high evolutionary sequence conservation. Residues from the conserved C4HC3 signature are marked in blue (see also Figure S5A and Data S1 sheet “DPF2_orthologs”). Numbers I1–I5 indicate the individual with the respective variant. Positions with de novo missense variants are indicated with a red arrow. Gray shading represents conservation.
(D) Amino acid sequence alignment of DPF2 and putative human paralog proteins with similar tandem PHD fingers shows conservation of the C4HC3 signature (see also Figure S5B and Data S1 sheet “PHD_finger_proteins_(PHF)”). Protein sequences were obtained from NCBI and ClustalW,21 and the msa package22 within R was used for alignment. References for the orthologs are as follows: H. sapiens (DPF2), GenBank: NP_006259.1; M. mulatta (LOC721967), GenBank: XP_002808108.1; M. musculus (Dpf2), GenBank: NP_035392.1; G. gallus (DPF2), GenBank: NP_989662.1; D. rerio (dpf2), GenBank: NP_001007153.1; and X. tropicalis (dpf2), GenBank: NP_001184101.1. References for the paralogs are as follows: DPF1, GenBank: NP_001128627.1; DPF2, GenBank: NP_006259.1; DPF3, GenBank: NP_001267471.1; KAT6A, GenBank: NP_006757.2; KAT6B, GenBank: NP_036462.2; and PHF10, GenBank: NP_060758.2.