Figure 3.
Functional Consequences of the DPF2 Missense Variants on Histone Binding and Sub-nuclear Localization
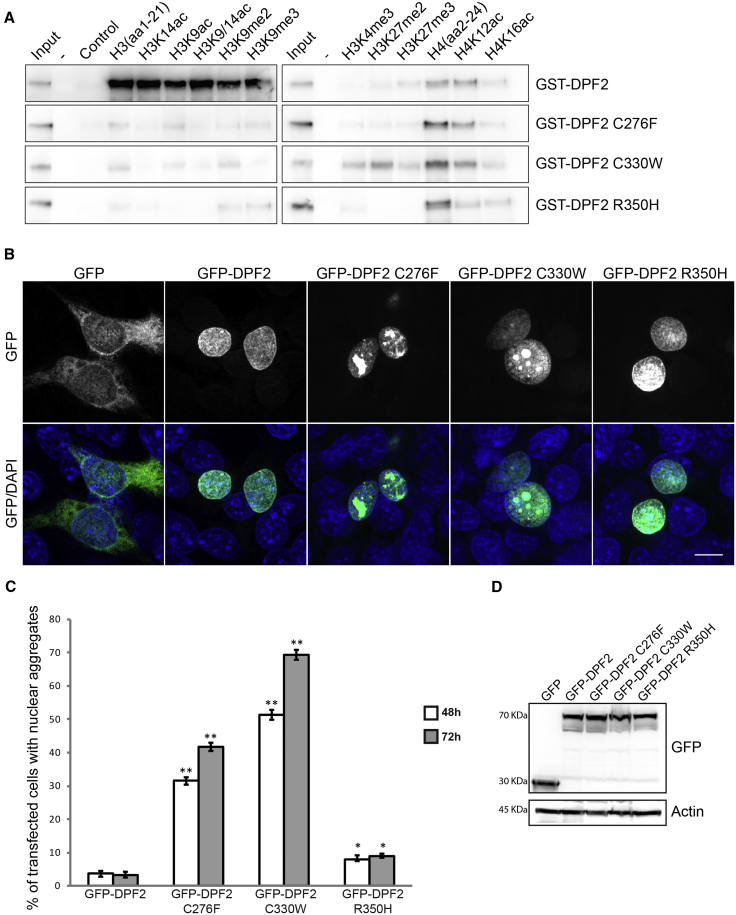
(A) Western blot analysis of histone peptide pull-downs shows the absence or residual binding of the GST-DPF2 recombinant proteins harboring the three missense variants to unmodified, acetylated, or methylated H3 peptides (wild-type GST-DPF2 is shown for comparison). Note that the interaction with unmodified and acetylated H4 peptides was not affected. Purified GST-fusion proteins and histone peptides are indicated. Input represents 5% of the purified GST proteins used in the pull-downs. As a control, no peptide was added in the pull-down assay. GST-tagged wild-type DPF2 and mutants were generated by N-terminal subcloning from the pEGFP-C3 into a pGEX4T1(GST) vector via BglII and SalI restriction sites (Table S4). Expression of GST-DPF2 fusion proteins in BL21 bacteria was induced with 0.1 mM IPTG. Collected cells were resuspended in 25 mM HEPES (pH 7.5), 500 mM KCl, 0.2 mM DTT, 1 mM EDTA, 10% glycerol, and proteinase inhibitor (Roche) and lysed with a French pressure cell press and then sonicated. Proteins were purified with Glutathione Sepharose 4B (GE Healthcare) according to the manufacturer’s instructions and eluted with 10 mM glutathione (pH 8.5). The eluted protein was dialysed with the Slide-A-Lyzer MINI Dialysis Device (ThermoFisher Scientific) against 25 mM HEPES (pH 7.5), 500 mM KCl, 0.2 mM DTT, 1 mM EDTA, and 10% glycerol for 2 hr and the same buffer with 60% glycerol overnight. Histone-peptide binding assays were performed as described in Lange et al.41 Biotinylated histone peptides were purchased from Millipore (unmodified H3, 12-403; H3K9ac, 12-431; H3K14ac, 12-425; H3K9/14ac, 12-402; H3K9me2, 12-430; H3k9me3, 12-568; H3K4me3, 12-460, H3K27me2, 12-566; H3K27me3, 12-565; unmodified H4, 12-372) and Tebu-bio (H4K12ac, 12-0032; H4K16ac, 12-0033).
(B) Representative confocal immunofluorescence microscopy images of HEK293 cells overexpressing empty GFP vector (column 1) and wild-type DPF2 (column 2) and mutant (columns 3–5) GFP-tagged constructs. GFP was homogeneously distributed throughout the cell, whereas wild-type DPF2 and mutants exhibited exclusively nuclear localization. Note that C276F and C330W mutants led to the formation of nuclear protein aggregates, whereas R350H had a minor effect. The top panel shows GFP, and the lower panel shows merged pictures with DAPI. Scale bar, 10 μm. Wild-type DPF2 cDNA was amplified by PCR. The product was N-terminally inserted into the pEGFP-C3 vector with the In-Fusion HD Cloning Plus Kit (Clontech, Takara) according to the manufacturer’s instructions. The variants in DPF2 were generated by mutagenesis PCR. Primers used for cloning and mutagenesis are described in Table S4. HEK293 cells were grown as described in Vasileiou et al.42 3 × 105 HEK293 cells were cultured on coverslips and transfected with 1 μg GFP and GFP-fusion plasmids with the use of polyethylenimine. After 48 hr, cells were fixed with 100% methanol at −20°C and stained with DAPI. Fluorescence images were acquired on an LSM 800 confocal laser scanning microscope (Carl Zeiss) with a 63× lens.
(C) HEK293 cells were examined 48 and 72 hr after transfection, and graphs indicate the percentage of cells exhibiting nuclear aggregates. Error bars represent the mean ± SEM, and significant values are indicated with ∗p < 0.05 or ∗∗p < 0.01 (unpaired Student’s t test). 100 random cells were used per experiment (n = 3).
(D) Western blot analysis from lysates of HEK293 cells transfected with the indicated plasmids with the use of anti-GPF (top) and anti-panActin (bottom). Molecular weight (kDa) is indicated on the left of the western blotting panels.