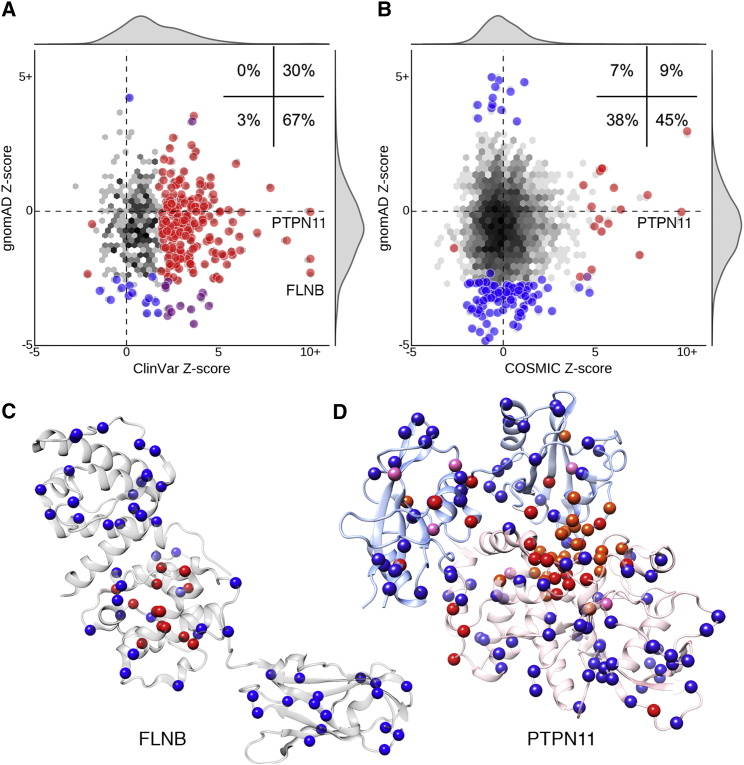
Figure 3.
Pathogenic and Neutral Missense Variants Have Distinct Spatial Distributions
(A and B) Comparison of the gnomAD missense Z-scores against ClinVar pathogenic (A) and COSMIC recurrent somatic (B) univariate Z-scores for experimentally derived protein structures. The inset reports the percentage of significant structures in each quadrant. The distribution over all structures is shown as a density plot, with black indicating higher density (log-scale). Large circles indicate structures with significant spatial distributions of either set of variants (two-sided permutation p value, FDR < 10%). Circles are colored red if the structure exhibits significant constraint on the variant set plotted on the x-axis, blue for significant contraint on the y-axis variant set, and purple if there is significant on both.
(C) Pathogenic variants (red) in FLNB (PDB: 4B7L) are clustered in the second calponin-homology domain, responsible for actin binding; neutral variants (blue) are distributed throughout the structure.
(D) Germline disease-causing (red) and recurrent somatic (pink) missense variants in PTPN11 (PDB: 5I6V) are clustered and frequently overlapping (orange) at the structural interface of the PTP (pink ribbon) and SH2 (blue ribbon) domains.