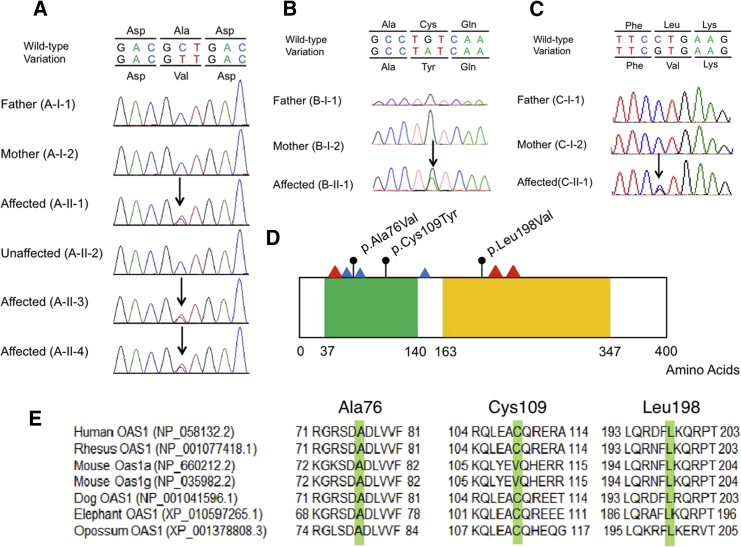
Figure 1.
OAS1 Variants in Three Families with PAP and Evolutionary Conservation and Variations in OAS1
(A–C) Genetic analysis for OAS1 variation. Sanger sequencing demonstrated variations of (A) c.227C>T (p.Ala76Val) in family A, (B) c.326G>A (p.Cys109Tyr) in family B, and (C) c.592C>G (p.Leu198Val) in family C. Black arrows show the positions of the variations.
(D) Schematic representation of OAS1 mutations. Black circles indicate the variants identified in this study. Blue and red triangles indicate metal binding sites (Asp75, Asp77, and Asp148) and ATP binding sites (Ser63, Lys213, and Gln229), respectively. Green and yellow boxes show the nucleotidyltransferase (NTP_transf_2) domain and 2′,5′-oligoadenylate synthetase 1, domain 2, and C terminus (OAS1_C) domain, respectively, predicted using the SMART program (see Web Resources).
(E) Evolutionary conservation of Ala76, Cys109, and Leu198 amino acids in human OAS1. These amino acids are highlighted in green. Six orthologous sequences were aligned using the multiple sequence alignment program, Clustal Omega (see Web Resources).