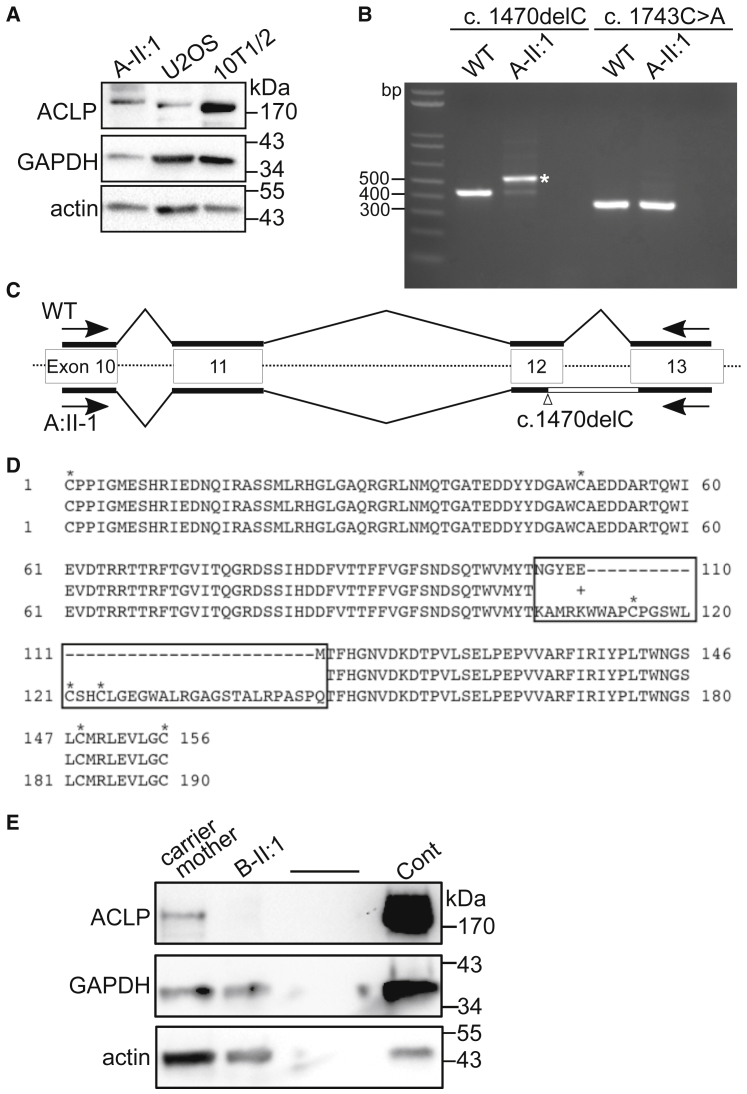
Figure 3.
Analysis of AEBP1 Mutant Fibroblast mRNA and Protein
Fibroblasts were isolated from dermal skin biopsies. Cells were routinely cultured in Dulbecco’s Modified Eagle’s Medium (Corning) with 3.7 g/L glucose, 10% fetal bovine serum (Hyclone), and 1% penicillin/streptomycin at 37°C in an 5% CO2 incubator. Cells were passaged before they reached confluence using 0.25% trypsin/EDTA. All cells were genotyped by PCR of genomic DNA and sequencing prior to further analysis.
(A) Western blot analysis of protein lysates derived from fibroblasts from subject A-II:1. Anti-ACLP antibodies detected a band >170 kDa. Protein extracts were prepared as previously described14, 18 by washing the cells in cold PBS followed by extraction in 25 mM Tris (pH 7.4), 50 mM sodium chloride, 0.5% sodium deoxycholate, 2% NP-40, and 0.2% sodium dodecyl sulfate (SDS) with 1× Complete protease inhibitors cocktail (Roche) and 1× PhosStop phosphatase inhibitors cocktail (Roche). Lysates were incubated on ice for 15 min and cleared by centrifugation at 12,000 × g for 10 min at 4°C. The supernatant was collected and protein concentration was measured using the BCA kit (Thermo Scientific). Protein aliquots were run on 4%–20% Novex SDS-polyacrylamide gels and transferred to nitrocellulose. Blots were probed with antibodies against ACLP14 and normalized to GAPDH (Sigma G9545, 1:10,000) and pan-actin (Thermo MS-1295B, 1:1,500).
(B) Fibroblasts were cultured in 12-well plates and total RNA was isolated and purified using GeneJET RNA Purification Kit (Thermo Scientific). cDNA was obtained using Maxima Reverse Transcriptase (Thermo Scientific). Regions of interest were amplified by PCR using OneTaq DNA polymerase (New England Biolabs) and the following primers: c.1470delC 5′-CCCATTGGGATGGAGTCACA-3′ (forward) 5′-CAGGTGAGTGGGTAGATGCG-3′ (reverse) producing a 422 bp product, and c.1743C>A 5′-GGCTCGTTTCATCCGCATCTA-3′ (forward) 5′-GCACCTCGTTGCCATGGAT-3′ (reverse) producing a 341 bp product. PCR products and controls were run on 2% agarose gels and bands of interest were purified using the QiaQuick Gel Extraction Kit (QIAGEN) followed by sequencing (Eton Bioscience). RNA was extracted from human ACLP-wild-type fibroblasts and from subject A-II:1, and following reverse transcription was amplified with PCR using primers spanning both genomic mutations. A band of approximately 520 bases was amplified (indicated by an asterisk).
(C) The DNA fragment (asterisk in B) was purified and subjected to DNA sequencing, which revealed that after the deletion in exon 12, the next intron was retained in the cDNA.
(D) The predicted amino acid sequence generated from the results of DNA sequencing and alignment with the wild-type sequence. The boxed region highlights different and additional amino acids, including the addition of three cysteines (indicated by an asterisk).
(E) Western blot analysis of protein lysates derived from fibroblasts from subject B-II:1 and his carrier mother with a murine fibroblast control.