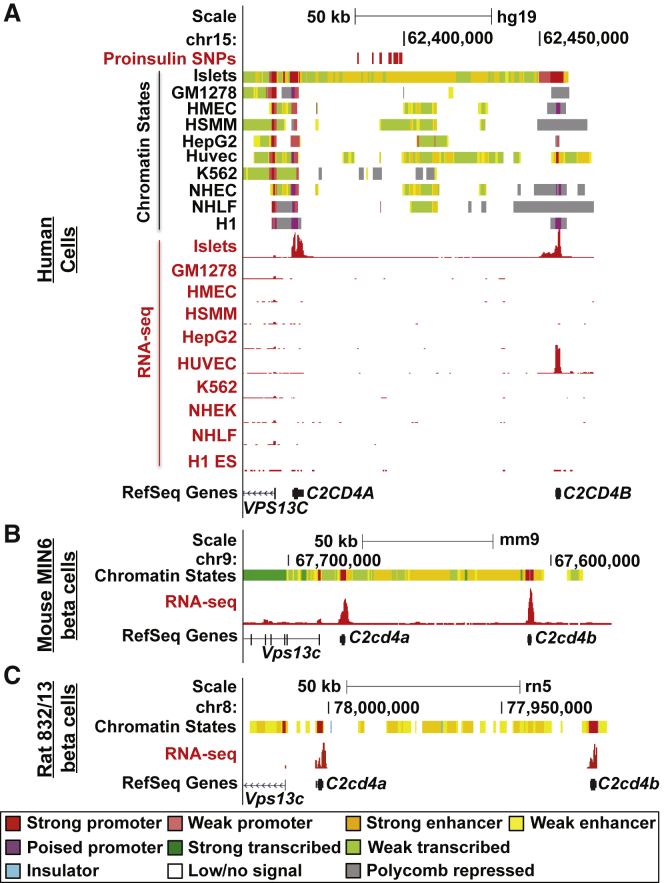
Figure 2.
Fine-Mapped Variants Reside in an Evolutionarily Conserved Islet Stretch Enhancer
UCSC Genome Browser views of chromatin states (upper panels) and RNA-seq (lower panels, red) in (A) human pancreatic islets and nine ENCODE cell types (GM12878, lymphoblastoid cells; HMECs, human mammary epithelial cells; HSMMs, human smooth muscle myoblasts; HepG2, hepatocellular carcinoma cells; HUVECs, human umbilical vein endothelial cells; K562, erythroleukemia cells; NHEKs, normal human epidermal keratinocytes; NHLFs, normal human lung fibroblasts; and H1 ES, embryonic stem cells), (B) mouse MIN6 β cells, and (C) rat INS-1(832/13) beta cells. RefSeq Genes indicate the location of C2CD4A, C2CD4B, and VPS13C in the region. Chromatin-state colors are as indicated. Increased low or no-signal states in 832/13 are due to low mappability. 16 proinsulin-associated variants are indicated in (A) (upper panel in red). UCSC Genome Browser coordinates correspond to hg19, mm9, or rn5.