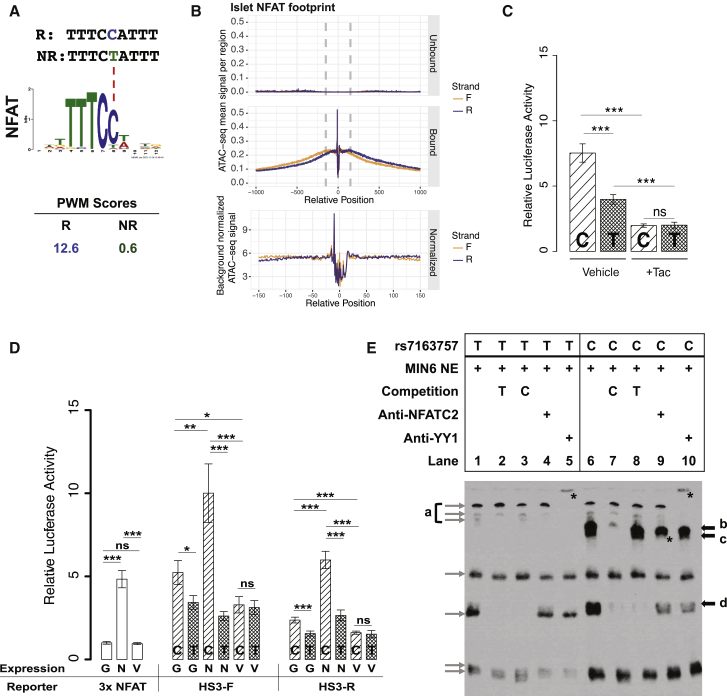
Figure 6.
NFAT Modulates Enhancer Activity of the rs7163757 Risk Allele (C)
(A) PWM scores (HaploReg) identify NFAT as the most plausible TF binding to the risk allele (C), but not to the non-risk allele (T), of rs7163757. Sequence is shown for the risk allele (R, blue) and the non-risk allele (NR, green). A dashed red line points to the affected position in the NFAT binding motif.
(B) Islet NFAT footprints. Density plots indicate normalized sequence coverage of ATAC-seq at the NFAT motifs predicted as bound (including the rs7163757 site) or unbound in two human islet samples.
(C) Luciferase reporter assay of HS3 (cloned in the forward orientation) containing the rs7163757 risk allele (C) or non-risk allele (T) in MIN6 cells treated with 10 ng/mL tacrolimus (Tac) or ethanol control (vehicle). Four to five biological replicates were measured in three independent experiments. Luciferase activity of HS3 constructs were normalized to that of the corresponding vehicle-control- and tacrolimus-treated pGL4.23 empty vector. Error bars represent mean luciferase activity ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (unpaired two-sided t test).
(D) Luciferase reporter assay of HS3 (cloned in forward and reverse orientations) containing the rs7163757 risk allele (C) or non-risk allele (T) in the presence of EGFPC1-huNFATc1EE-WT or the NFAT inhibitor construct EGFPN1-VIVIT. pGL3-NFAT-Luciferase (NFAT reporter), containing three canonical NFAT binding sites upstream of a luciferase reporter, plus EGFP plasmid served as a control for NFAT activity of the expression constructs. For all cloned HS3 enhancer sequences co-transfected with EGFP, EGFPC1-huNFATc1EE-WT, or EGFPN1-VIVIT, eight biological replicates were measured in four independent experiments. Luciferase activity of NFAT expression constructs was normalized to the NFAT reporter plus EGFP. Luciferase activity of HS3 expression constructs with EGFP or NFAT expression constructs was normalized to that of the pGL4.23 empty vector with EGFP or NFAT expression constructs, respectively. HS3-F and HS3-R denote the HS3 sequence (C or T allele) cloned into pGL4.23 in the forward and reverse orientations, respectively; G, EGFP; N, EGFPC1-huNFATc1EE-WT; and V, EGFPN1-VIVIT. Error bars represent mean luciferase activity ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (unpaired two-sided t test).
(E) Allele-specific protein binding of sequences containing different rs7163757 alleles. EMSA with biotin-labeled probes containing the rs7163757 T or C allele incubated with MIN6 nuclear extract (NE). Black arrows indicate allelic differences in protein binding. Gray arrows show non-specific binding or no clear differences between alleles. Bands labeled b, c, and d indicate proteins that bind specifically to the C allele. The asterisk in lane 9 indicates the disruption of band c by NFATc2 antibody. Asterisks in lanes 5 and 10 indicate supershift of non-allele specific band a by YY1 antibody.