Figure 1.
Generation of the Otud7a Knockout Mouse Model
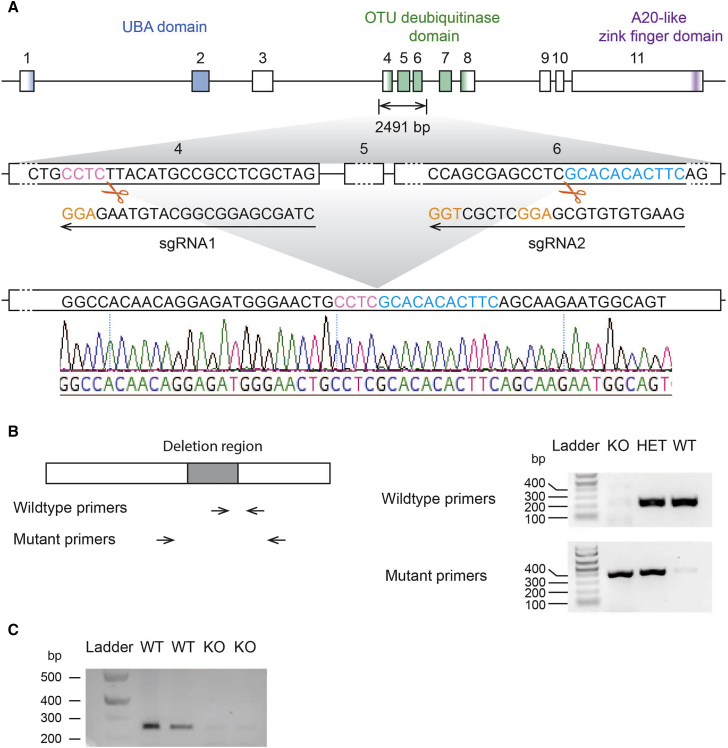
(A) Scheme of Otud7a knockout design and sequencing validation. Exons of Otud7a are boxed, whereas introns are represented by lines in between exons. All exons and introns are plotted in scale within their category. Three domains have been identified in OTUD7A: the UBA domain, OTU deubiquitinase, and an A20-like zinc finger domain. The nucleotides encoding these domains are colored blue, green, and purple, respectively. NGG sequences in the two sgRNAs are shown in orange. Following CRISPR/Cas9 manipulation, we obtained mice with knockout of the expected region, bordering sequences of which are highlighted in pink and blue. Sequencing trace from tail DNA of the mice are shown at the bottom.
(B) Genotype identification of OTUD7A-deficient mice. The knockout region was boxed in gray. Genotyping primer binding regions for wild-type and knockout primer sets were indicated by arrows. In the representative genotyping results, animals that carry a wild-type allele show bands at 223 bp with wild-type primers, and those that carry a knockout allele show bands at 389 bp with mutant primers.
(C) RT-PCR products with RNA extracted from brains of wild-type and Otud7a knockout mice showing a deficiency of Otud7a transcripts in the knockout animals. Primers used for RT-PCR bind to exon 3 and 8 respectively. Forward primer: 5′-GGACTTCAGGAGCTTCATCG-3′, reverse primer: 5′-ACCTCCAAGGGCAGGTAGAT-3′. Expected PCR product size is 244 bp.