Abstract
Phosphatidic acid (PA) is an important signaling lipid that plays roles in a range of biological processes including both physiological and pathophysiological events. PA is one of a number of signaling lipids that can act as site-specific ligands for protein receptors in binding events that enforce membrane-association and generally regulate both receptor function and subcellular localization. However, elucidation of the full scope of PA activities has proven problematic, primarily due to the lack of a consensus sequence among PA-binding receptors. Thus, experimental approaches, such as those employing lipid probes, are necessary for characterizing interactions at the molecular level. Herein, we describe an efficient modular approach to the synthesis of a range of PA probes that employs a late stage introduction of reporter groups. This strategy was exploited in the synthesis of PA probes bearing fluorescent and photoaffinity tags as well as a bifunctional probe containing both a photoaffinity moiety and an azide as a secondary handle for purification purposes. To discern the ability of these PA analogues to mimic the natural lipid in protein binding properties, each compound was incorporated into vesicles for binding studies using a known PA receptor, the C2 domain of PKCα. In these studies, each compound exhibited binding properties that were comparable to those of synthetic PA, indicating their viability as probes for effectively studying the activities of PA in cellular processes.
Introduction
Binding interactions in which receptors, termed peripheral proteins, interact with the surfaces of cellular membranes are known to control a litany of crucial physiological and pathophysiological processes. While a number of mechanisms for membrane targeting of cytosolic proteins exist, a common motif involves the actions of lipids such as phosphatidic acid (PA),1–3 diacylglycerol (DAG),4,5 and the phosphoinositides (PIPns)6–11 as site-specific ligands that enforce the membrane-anchoring of protein receptors. The presence of signaling lipids in cellular membranes is tightly regulated both spatially and temporally, and thus their presentation directly controls the localization of receptors within the cell.12 In addition, the binding of peripheral proteins to the membrane generally regulates the function of the receptor. Protein function can either be directly affected by lipid binding or indirectly regulated, such as through interaction with another protein that is localized to the membrane surface.4,5,13,14 It is now apparent that protein–lipid binding events are particularly prominent in regulating crucial processes, and thus defects in binding and lipid composition result in serious diseases. For example, despite the relative simplicity of its structure, PA plays a vital role in carcinogenesis by regulating the function of Raf-1 kinase through binding, a protein that is involved in vital cellular pathways.15,16 As a result of these implications, it is important to elucidate the complex details pertaining to protein–lipid binding interactions.
To date, a number of receptors that target PA2,3 have been identified, including protein kinases,17–19 protein phosphatases,20–22 and cAMP-specific phosphodiesterases.23,24 A particularly well studied example is Raf-1 kinase, an enzyme that interacts with cellular membranes by binding both PA and phosphatidylserine (PS) using separate binding domains.25 When bound to the membrane surface, Raf-1 forms a protein–protein interaction with membrane-associated Ras GTPases, which activates Raf-1 to initiate the Ras/Raf/MEK/ERK cell proliferation pathway that is aberrant in cancer.25–32 Other receptors that interact with PA include certain isoforms of the protein kinase C (PKC) family, such as PKCα33 and PKCε,34 which are heavily involved in cancer onset. This family consists of at least eleven isozymes that exhibit varying lipid-binding properties, opposing or overlapping functions, and differing localization profiles, complicating their study.4,5,35–38 Each PKC contains two C1 domains that interact with DAG as well as a C2 domain that often binds to a phospholipid target that varies among the different isoforms, including PA and PS. The exact roles of the different PKCs in carcinogenesis remain unknown due to challenges in characterizing variations in activity that occur among the isozymes.39–41 A current hypothesis is that subtle differences in lipid-binding patterns between the PKC isozymes result in discrepancies in their subcellular localization that in turn regulate function.5,42
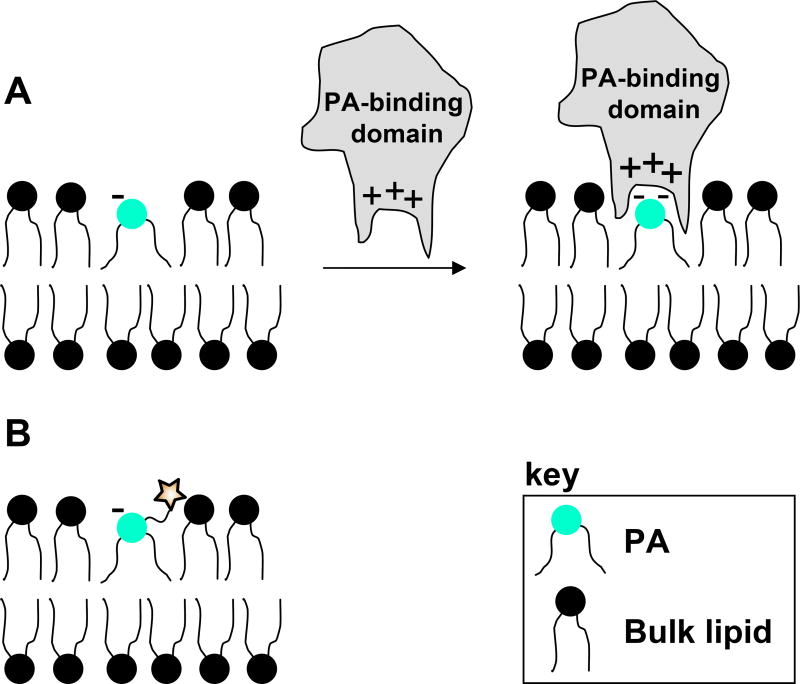
One focus of research seeks to understand how receptors could possess specificity in the binding of PA over other lipids bearing negative charge that are often significantly more abundant. Problems related to this issue often lead to the misidentification of PA receptors.2,43 de Kruijff and coworkers have presented an electrostatic hydrogen bond switch mechanism to explain the specificity of PA as a ligand despite its simple structure.43,44 Their results showed that monoprotonated PA is stabilized by an intramolecular hydrogen bond that preserves the -1 charge, causing proteins and peptides with basic residues to associate to the membrane via electrostatic attraction. Following this initial interaction, hydrogen bonds are formed between the receptor and PA headgroup that immediately lead to full phosphate deprotonation (-2 charge). This charge increase enhances association and provides binding specificity. The shape of PA is also believed to contribute to binding specificity.43 At physiological pH, PA exists as a cone-shaped structure, which causes the phosphate headgroup to lie below the membrane surface in the mildly polar interfacial region.45 This presentation is thought to enhance the penetration of hydrophobic residues into the membrane around the PA binding domain. The presentation of PA in the membrane and the charge-reinforced H-bonding enforced during binding are depicted in Figure 1A. Both of these concepts are observed in the binding of PA by Raf-1 kinase,25,27,43 and PKCs,33,34 which contain basic amino acids that are crucial for PA recognition as well as hydrophobic residues that penetrate the membrane during binding.
Figure 1. PA membrane presentation and protein binding.
A. PA presentation in the membrane and charge reinforced H-bond formed during protein binding. Figure is adapted from reference 43. B. Expected presentation of tagged PA probes in the membrane.
Despite these advancements in understanding the actions of PA as a ligand, the full scope of PA-targeting proteins is not yet clear as the number of identified receptors is relatively small compared to the biological processes that show PA-dependence. A primary complication in the discovery of PA-binding proteins is that these receptors generally don’t possess conserved consensus sequences that can be employed for sequence homology searching.2,3,8 The PA binding domains established to date are diverse and consist of sequences that were not previously known to bind lipids.2 In fact, the only common link that many of the PA binding domains share are small clusters of basic (Arg, Lys, Trp) amino acids.2,3,8,43 Due to this inability to discover PA-binding receptors via structure searches, experimental approaches are crucial for elucidating binding interactions. As such, chemical probes are vital for studying and characterizing protein–PA binding interactions at the molecular level.
Results and Discussion
Synthetic PA probes have previously proven effective for probing receptors that target this lipid. For example, Nemoz and coworkers developed a photoaffinity-tagged PA probe that was used in the covalent labeling of type 4 cAMP-phosphodiesterases (PDE4).46 The photoaffinity label was incorporated into the acyl tails of commercial lipids, which is effective for proteins that interact significantly with lipid chains buried within the hydrophobic membrane core. Ktistakis and coworkers have designed a fully synthetic PA probe that was attached to beads via an amine incorporated into the sn-1 acyl tail.47,48 The resulting PA-functionalized resin was employed for affinity chromatography to selectively purify receptors that target this lipid. In addition, Prestwich and coworkers have developed synthetic fluoromethylene phosphonate analogs of PA as stabilized derivatives that are capable of activating the mTOR pathway.49 Gadella and coworkers developed a caged PA analog that was used to induced flagellar excision in algae.50 The introduction of an NPE caging moiety onto commercial lipids allowed the probe to cross the membrane, and photolysis produced an increase of PA within the cell that decreased the swimming of the algae. Finally, Schultz and coworkers have recently employed a bifunctional PA probe with a hydrolysable phosphate protecting group to achieve in vivo labeling of PA via bioorthogonal copper-free click chemistry.51
While these studies have proven lipid probes to be effective for characterizing biological activities, much remains to be learned about the roles of PA in cellular processes, and thus further probe strategies are necessary. The synthesis of lipid probes from scratch is beneficial as this increases cost efficiency, facilitates purification of the final probes, and allows for the exploration of a range of different modification strategies. For example, introduction of reporter groups in place of a hydrogen at the sn-1 headgroup position, which has rarely been pursued,52 is expected be effective for placing the added group proximal to a bound protein but away from the phosphate headgroup, the primary recognition moiety. However, a significant impediment to achieving broadly applicable probe strategies is the synthesis required to produce each reporter-functionalized probe. Therefore, the optimal probe development approach would provide modularity such that a diverse array of reporter groups could be conveniently introduced at a late stage in the synthesis to access numerous probe structures.
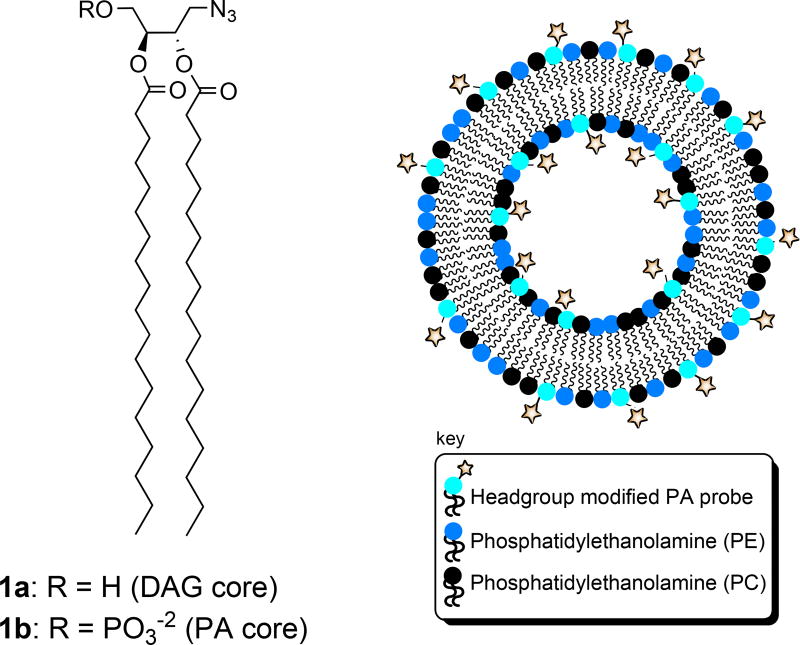
Recently, we reported a modular approach to lipid probe synthesis that was employed for the synthesis of a range of DAG probes that each possess a reporter tag in place of a hydrogen at the headgroup sn-1 position.53 Functionalized probes were obtained from a core DAG scaffold bearing an azidomethyl group at the sn-1 position (1a, Figure 2) via the bioorthoganol azide-alkyne cycloaddition (click chemistry) reaction.54–56 Similar strategies have recently proven effective for facile derivatization of lipid scaffolds for bolaamphiphile development,57 and delivery and imaging58 applications. Despite modification of the headgroup position of DAG, the resulting probes exhibited Kd values in the binding of PKCδ that were virtually identical to natural DAG, indicating that this is a viable derivatization strategy.
Figure 2. Modular azide-tagged scaffolds corresponding to DAG and PA and expected presentation in liposomes employed for binding studies.
Herein, we describe the extension of this probe strategy for the development of biologically active PA probes. Our design for a modular PA scaffold (1b) builds upon the success of the previous DAG analogue (1a). Initially, we attempted to synthesize 1b by simply installing the phosphate headgroup onto DAG scaffold 1a. However, as was the case during a number of attempts at the synthesis of 1a,53 we found that the use of phosphoramidite chemistry for this purpose led to a significant amount of sn-2 to sn-3 acyl migration, and thus this approach was not viable. In order to avoid the problems associated with acyl migration in hydroxyl-containing glycerol intermediates, our focus shifted to the development of a route in which the phosphate headgroup and azide tag are introduced early in the synthesis, prior to installation of the lipid acyl chains.
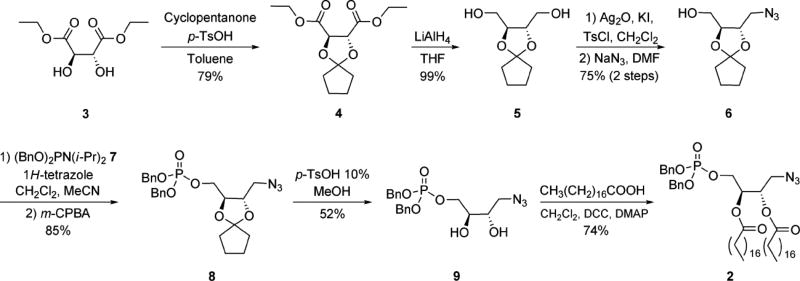
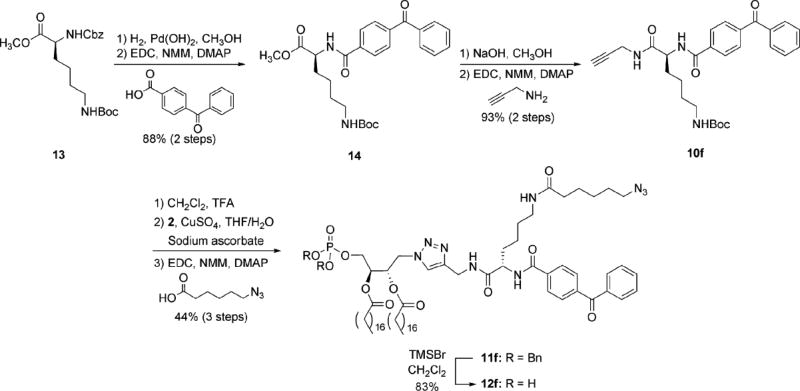
The current approach to the efficient development of numerous derivatized PA probes exploits protected azide-derivatized PA precursor 2 as a modular scaffold. The final synthetic route to this compound is indicated in Scheme 1. Protected tartrate analogue 5 was obtained from commercial diethyl-l-tartrate (3) using modified literature procedures for cyclopentylidene introduction to 4 and subsequent ester reduction. Next, mono-tosylation of bis-hydroxymethyl compound 5 was followed by azide substitution to furnish 6. The phosphate headgroup, protected as a phosphotriester, was then installed using dibenzyl diisopropylphosphoramidite (7) to afford 8. Careful removal of the cyclopentylidene acetal under acidic conditions was then employed to access diol 9. Finally, the acyl chains were introduced using traditional coupling reagents for esterification to obtain dibenzyl-protected azido-PA derivative 2. A key aspect of this synthesis was the determination of the optimal protecting group for the interior diol that could be removed to afford 9 without affecting the phosphotriester and azide groups. While several protecting group strategies were explored (see Supporting Information for details), the cyclopentylidene group of 7 proved most effective.
Scheme 1. Synthetic route to protected azide-tagged modular PA scaffold 2.
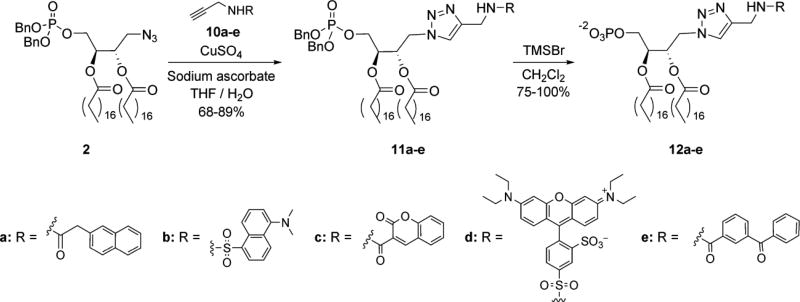
Following the synthesis of phosphotriester-protected PA analogue 2, we employed this scaffold for the efficient production of a number of derivatized PA probes (Scheme 2). The optimal approach involved initial reporter group introduction using click chemistry to couple scaffold 2 to a range of functionalized alkynes (10a–e)53 in the generation of phosphotriesters 11a-e. The phosphotriester groups were subsequently deprotected with bromotrimethylsilane to generate PA probes 12a-e. This strategy was exploited to obtain PA probes derivatized with a range of fluorescent dyes (naphthyl (12a), dansyl (12b), coumarin (12c), and rhodamine (12d)) as well as with a photoaffinity tag (benzophenone (12e). Among the fluorescent analogues are different Förster resonance energy transfer (FRET) pairs, which are useful for detecting changes in proximity of donor and accepter-tagged molecules.59–63 Installation of the photoaffinity tag is advantageous for photo-cross-linking studies64 to characterize binding and map the location of binding domains on protein receptors.
Scheme 2. Convenient derivatization of protected PA scaffold 2 for the efficient production of derivatized PA probes 12a–e.
In addition to probes 12a–e, we also sought to produce a bifunctional PA analogue that could be employed to purify, identify, and characterize PA-binding receptors. A significant challenge that has hindered elucidation of the full scope of PA activities is the difficulty associated with identifying PA-binding receptors, primarily due to the lack of a consensus sequence. Our strategy for probe development is inspired by the concept of activity-based protein profiling (ABPP), which uses substrate analogues as probes for a mechanism-based approach to the collective labeling of enzyme targets based on their activity.65–67 In the current approach, a PA probe (12f, Scheme 3) bearing both a photo-cross-linking group (benzophenone) for attachment to cognate receptors, as well as a secondary tag (azide) for purification of successfully cross-linked proteins is designed to fish out PA-binding receptors from a complex sample. Similar bifunctional designs have previously proven effective for this purpose,68,69 including a recent study that utilized a PC probe to identify receptors that target this lipid.70
Scheme 3. Application of modular scaffold 2 for the production of bifunctional PA probe 12f.
The design of probe 12f employs a lysine moiety as a Y-shaped linker to introduce both the photo-cross-linking and azide groups onto PA. Thus, the synthesis of probe 12f commenced with fully protected lysine 13. First, hydrogenolysis was used to remove the carbobenzyloxy (cbz) group, followed by coupling to 4-benzoylbenzoic acid to introduce the benzophenone moiety of 14. Methyl ester hydrolysis was then performed prior to coupling with propargylamine in the generation of alkyne 10f. Next, the tert-butoxycarbonyl (boc) group of 10f was removed, followed by the use of click chemistry to introduce PA scaffold 2, and finally coupling of the newly deprotected amine with 6-azidohexanoic acid71 to produce phosphotriester-protected PA analogue 11f. This was then deprotected with bromotrimethylsilane as before to access bifunctional probe 12f.
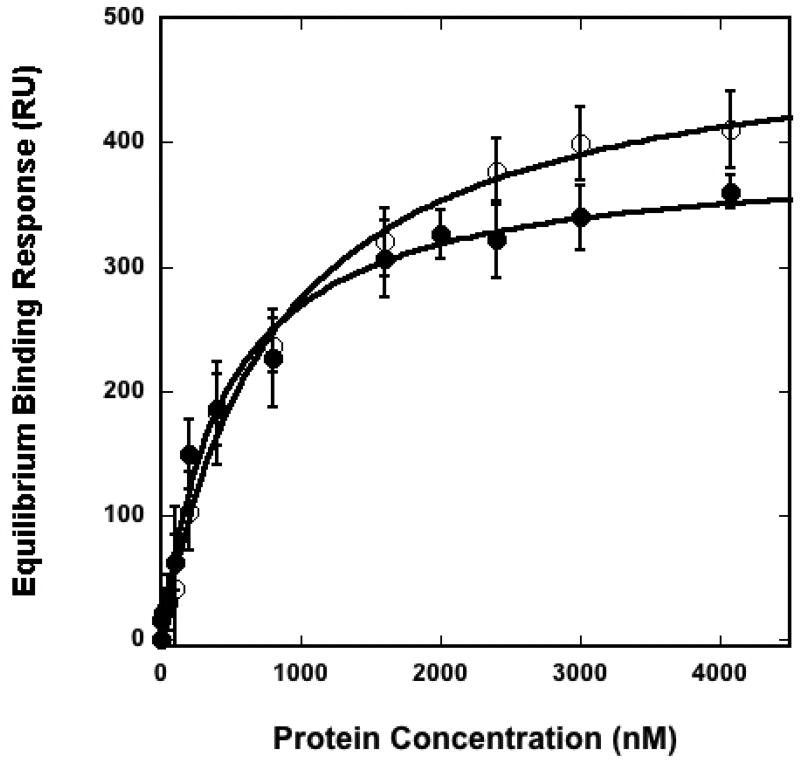
With these seven new derivatized PA probes (12a–f) that resulted from our modular approach in hand, we sought to determine the extent to which each compound retained the typical binding properties of PA effectors. To do so, we screened the binding of the C2 domain of PKCα to lipid vesicles containing the different PA analogues at 20 mol %. Studies employed an isolated C2 binding domain to increase the specificity for PA since full length PKCα contains multiple binding domains that target different lipids and assist in membrane association. For binding studies, a surface plasmon resonance (SPR) assay was implemented using POPC/POPE/PA (40:40:20) vesicles containing the various PA probes as an active surface (see depiction of liposome composition in Figure 2). We first performed binding experiments with vesicles containing synthetic unmodified POPA, as the binding of PKCα-C2 to PA-containing vesicles has been well documented.72 In this study, the binding of PKCα-C2 to these vesicles yielded a Kd value of 440 ± 50 nM, as shown in Table 1. Next, we systematically analyzed the binding efficacy of the described PA reporters. The resulting binding affinities for each compound are indicated Table 1. In addition, an overlay of representative binding curves corresponding to vesicles containing synthetic PA (POPA) and dansyl-PA (12b) is shown in Figure 3, and raw SPR sensorgrams are shown in supplementary figure S51.
Table 1.
Dissociation constants measured for binding of PKCα–C2 to vesicles containing PA derivatives.
| POPC:POPE:PA (40:40:20) | Kd (nM)a |
|---|---|
| Synthetic PA (POPA) | 440 ± 50 |
| Azido PA (1b) | 640 ± 140 |
| Naphthyl PA (12a) | 780 ± 50 |
| Dansyl PA (12b) | 800 ± 70 |
| Coumarin PA (12c) | 850 ± 120 |
| Rhodamine PA (12d) | 510 ± 90 |
| Benzophenone PA (12e) | 570 ± 150 |
| Benzophenone/Azido PA (12f) | 520 ± 80 |
| POPC:POPE:PA (55:40:5) | Kd (nM)a |
| Synthetic PA (POPA) | 490 ± 90 |
| Azido PA (1b) | 730 ± 130 |
| Coumarin PA (12c) | 940 ± 120 |
| Rhodamine PA (12d) | 810 ± 80 |
Binding experiments were performed in 25 mM Tris HCl pH 7.4, 150 mM NaCl, 1 mM CaCl2 and 1 mM DTT
Figure 3. Representative binding curves for the binding of PKCα-C2 to membranes containing different PAs.
SPR measurements were used to determine Kds of PKCα-C2 for POPC/POPE/PA (40:40:20) vesicles where PA represents synthetic PAs reported in this manuscript. PKCα-C2 was injected at varying concentrations to generate a binding isotherm of resonance units bound (saturating response units) versus protein concentrations. The solid lines represent a theoretical curve obtained based on Rmax and Kd values determined by nonlinear least squares analysis. Representative curves are shown for synthetic-PA (●) and dansyl-PA (○). Error bars are indicative of the standard deviation calculated from three separate measurements.
The results from these membrane binding studies indicate that each of the described PA probes displayed similar properties to POPA in the binding of PKCα-C2, with Kd values ranging from 510 to 850 nM. PA probes derivatized with rhodamine (12d, 510 nM), the bifunctional benzophenone/azide tag (12f, 520 nM) and benzophenone (12e, 570 nM), yielded the strongest binding affinities, which were comparable to POPA (440 nM). While the others, napthyl PA (12a, 780 nM) dansyl PA (12b, 800 nM), and coumarin PA (12c, 850 nM) yielded slightly weaker affinities, all are less than two times that of POPA. With these results, there doesn’t appear to be a clear trend that relates binding affinities to the physiochemical properties of the appended reporter groups.
These studies employing 20 mol% PA in liposomes proved useful in demonstrating the reliability of our synthetic PA probes, however, the physiological concentrations of PA in cellular membranes are thought to be much lower. To validate the significance and physiological relevance of PA binding, we monitored the interaction of PKCα-C2 with vesicles containing 5 mol% (POPC:POPE:PA 55:40:5) of the different PA analogues. As shown in Table 1, PKCα-C2 bound synthetic PA and corresponding PA probes with nearly the same affinity. Moreover, PKCα-C2 affinity for vesicles containing 5 mol% PA probes was comparable to that for vesicles containing 20 mol% PA probes, suggesting PKCα-C2 binding is saturated at 5 mol% PA. Future studies will be performed to further understand the presentation of these compounds in a membrane environment and the effect on protein binding properties.
Conclusion
Herein, we describe an efficient approach for the generation of a range of derivatized PA probes through modular functionalization of core azide-tagged scaffold 2 via click chemistry. Despite the fact that bulky reporter moieties are introduced onto the PA headgroup using this approach, the first quantitative analysis of these compounds indicates their ability to mimic PA in the recruitment of effectors to membrane surfaces. These studies demonstrate the relevance of these PA probes for use as robust reporters of PA activity in biological processes. Furthermore, the installation of reporter moieties at the lipid headgroup is expected to place the appended tag in proximity to proteins bound to the membrane surface (see schematic depiction of expected tag presentation in Figure 1B). The use of click chemistry is also beneficial in this regard, as the polar triazoles that result, when presented on lipids, are believed to preferentially localize to the aqueous environment rather than the hydrophobic membrane core.57 As a result, the triazole is anticipated to enhance presentation of the appended tag at the hydrophilic membrane surface. Headgroup modification, in cases where the perturbation of protein binding is limited, is expected to enhance challenging applications such as FRET-based detection of proteins bound to a tagged lipid, and protein–lipid photo-cross-linking, which require the tag to be in close proximity to a bound protein.
Currently, we are extending studies employing these PA probes in several ways. First, we are continuing the characterization of these compounds to further understand their efficacy in mimicking PA in terms of physiochemical properties, presentation in a membrane environment, and protein binding properties. In addition, we are applying these newly developed analogues as chemical tools to characterize the activities of PA via a range of approaches. These probes can be incorporated into liposomes as a model system for characterizing the details of protein–lipid binding events at the molecular level. Furthermore, the described probes can be modified for in vivo analysis of PA activities along the same lines as previous probe-based approaches. Probe 12f will be employed to purify and identify PA-binding receptors in similar fashion to recent reports.51 Finally, we are also advancing our modular probe strategy to develop biologically active analogues corresponding to other important signaling lipids.
Experimental
General
Generally, reagents were purchased from Acros or Aldrich and used as received. Cbz-Lys(Boc)-OMe was purchased from Chem-Impex International (Wood Dale, IL). Dry solvents were obtained from a Pure Solv solvent delivery system purchased from Innovative Technology, Inc. Column chromatography was performed using 230 – 400 mesh silica gel purchased from Sorbent Technologies. NMR spectra were obtained using a Bruker AC250 spectrometer updated with a TecMag data collection system, a Varian Mercury 300 spectrometer, and a Bruker Avance 400 spectrometer. Mass spectra were obtained with JEOL DART-AccuTOF spectrometer with high resolution capabilities. Optical rotation values were obtained using a Perkin-Elmer 241 polarimeter. HPLC data was obtained using an HP series 1100 HPLC with an Alltech Lichrosphere SI 60 5U column with HPLC grade solvents purchased from Fischer. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidic acid (POPA) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL).
(2R,3R)-diethyl 1,4-dioxaspiro[4.4]nonane-2,3-dicarboxylate (4)
Fully protected tartrate 4 was obtained following a procedure modified form that used to access the enantiomer of this structure.52 Diethyl-l-tartrate 3 (4.05 g, 19.6 mmol) was dissolved in toluene (130 mL). To this stirring solution was added cyclopentanone (8.7 mL, 98 mmol) and p-toluenesulfonic acid (373 mg, 1.96 mmol). A Dean–Stark trap was attached and the reaction was heated at 130 °C overnight. Solid sodium bicarbonate (329 mg, 20%) was added and stirring was continued for 10 min. The reaction mixture was filtered and the filtrate was condensed under reduced pressure. Column chromatography with silica gel and a gradient solvent system of 0–20% ethyl acetate to hexanes afforded 4 as a yellow oil (4.19 g, 79%).
Characterizations matched the enantiomer of 4 but for the change in sign of the optical rotation.52 [α]D296K – 31.5(c = 2.92, CHCl3); 1H NMR (300 MHz, CDCl3) δ 4.71 (s, 2H), 4.26 (q, J = 7.2 MHz, 4H), 2.03–1.67 (m, 8H), 1.30 (t, J = 7.2 MHz, 6H); 13C NMR (75.5 MHz, CDCl3) δ 169.7, 123.3, 77.0, 61.9, 36.6, 23.5, 14.1; HRMS [M + H]+ calcd: 273.13381, found: 273.13516.
(2S,3S)-1,4-dioxaspiro[4.4]nonane-2,3-diyldimethanol (5)
Protected tartrate 5 was obtained using a procedure modified from reports of the enantiomer52 and a similar compound.53 Diester 4 (2.5 g, 9.18 mmol), dissolved in anhydrous THF (8 mL), was added dropwise to a solution of lithium aluminum hydride (700 mg, 18.36 mmol) in anhydrous THF at 0 °C under nitrogen. After the addition was complete, stirring was continued for 1 h at 0 °C and then at room temperature for an addition 1 h. The solution was cooled to 0 °C and the nitrogen was removed. Carefully, water (1 mL), 10% NaOH (1 mL), and water (2 mL) were added to quench the reaction. Stirring was continued for 30 min at room temperature. Anhydrous magnesium sulfate was added and stirring was continued for 30 min. The solution was filtered and concentrated under reduced pressure. Column chromatography with silica gel and a gradient solvent system of 0–10% methanol to ethyl acetate afforded 5 as a clear oil which solidified upon refrigeration (1.70 g, 99%).
Characterizations matched the enantiomer of 5 but for the change in sign of the optical rotation.52 [α]D296K – 6.50 (c = 1.692, CHCl3); 1H NMR (300 MHz, CDCl3) δ 3.97–3.94 (m, 2H), 3.81–3.69 (m, 4H), 2.30–2.05 (m, 2H), 1.89–1.60 (m, 8H); 13C NMR (75.5 MHz, CDCl3) δ 119.4, 78.0, 62.3, 37.4, 23.4; HRMS [M + H]+ calcd: 189.11268, found: 189.11238.
((2S,3S)-3-azidomethyl)-1,4-dioxaspiro[4.4]nonan-2-yl)methanol (6)
Azide 6 was obtained following procedures reported for similar compounds.53,73 Diol 5 (720 mg, 3.83 mmol) was suspended in dichloromethane (20 mL). With stirring, silver (I) oxide (1.33 g, 5.75 mmol), tosyl chloride (803 mg, 4.21 mmol) and finely crushed potassium iodide (64 mg, 0.383 mmol) were added. The resulting solution was stirred at room temperature for 2 h and then filtered through a plug of silica gel with 100% ethyl acetate as the eluant. The filtrate was concentrated under reduced pressure, and then DMF (40 mL) and sodium azide (622 mg, 9.58 mmol) were added. The solution was stirred at 85 °C overnight, concentrated, extracted with chloroform (2 × 100 mL), dried with magnesium sulfate, and concentrated under reduced pressure. Column chromatography with silica gel and 50% ethyl acetate to hexane as eluant afforded 6 as a clear oil (636 mg, 78%).
[α]D296K – 57.8 (c = 1.492, CHCl3); 1H NMR (300 MHz, CDCl3) δ 4.05–3.90 (m, 2H), 3.81–3.62 (m, 2H), 3.55–3.51 (m, 1H), 3.30–3.33 (m, 1H), 2.65 (bs, 1H), 1.89–1.67 (m, 8H); 13C NMR (75.5 MHz, CDCl3) δ 119.8, 78.5, 76.1, 62.0, 52.0, 37.3, 37.2, 23.5, 23.3; HRMS [M – N2 + H]+ calcd: 186.11302, found: 186.11384.
((2S,3S)-3-(azidomethyl)-1,4-dioxaspiro[4.4]nonan-2-yl)methyl dibenzyl phosphate (8)
Azido alcohol 6 (425 mg, 1.99 mmol) was dissolved in anhydrous dichloromethane. 1H-tetrazole (13.3 mL, 5.98 mmol, 0.45 M stock) was added and the solution was cooled to 0 °C under nitrogen. Dibenzyldiisopropylphosphoramidite (7, 721 µL, 2.19 mmol) was then added dropwise. Stirring was continued for 10 min at 0 °C and then at room temperature for 1 h. At this point the reaction solution was cooled to 0 °C and m-chloroperoxybenzoic acid (1.81 g, 5.98 mmol, 57% purity) was added and stirring was continued for 1.5 h. The reaction was quenched with the addition of saturated sodium bicarbonate, extracted with dichloromethane (2 × 100 mL), dired with magnesium sulfate, and concentrated under reduced pressure. Column chromatography with silica gel and 50% ethyl acetate to hexane as eluant afforded 8 as a pale yellow oil (803 mg, 85%).
[α]D296K – 24.3 (c = 1.68, CHCl3); 1H NMR (300 MHz, CDCl3) δ 7.42–7.32 (m, 10H), 5.11–4.99 (m, 4H), 4.09–3.87 (m, 4H), 3.43–3.37 (m, 1H), 3.24–3.18 (m, 1H), 1.89–1.60 (m, 8H); 13C NMR (75.5 MHz, CDCl3) δ 135.7, 135.6, 128.7, 128.6, 128.0, 120.2, 76.8, 76.0, 75.9, 69.6, 69.5, 66.7, 66.6, 51.8, 37.3, 37.1, 23.5, 23.4; 31P NMR (121.5 MHz, CDCl3) δ 0.143; HRMS [M – N2 + H]+ calcd: 446.17325, found: 446.17276.
(2S,3S)-4-azido-2,3-dihydroxybutyl dibenzyl phosphate (9)
Phosphotriester 8 (1.07 g, 2.26 mmol) was dissolved in methanol (15 mL). With stirring was added p-toluenesulfonic acid (43 mg, 0.226 mmol). The solution was stirred at room temp overnight and quenched with saturated sodium bicarbonate and extracted with chloroform (2 × 100 mL), dried with magnesium sulfate, and concentrated under reduced pressure. Column chromatography with silica gel and a gradient solvent system of 80–100% ethyl acetate to hexanes afforded 9 as a clear oil (482 mg, 52%).
[α]D296K + 8.04 (c = 1.48, CHCl3); 1H NMR (300 MHz, CDCl3) δ 7.37–7.31 (m, 10H), 5.05–5.02 (m, 4H), 4.04–4.00 (m, 2H), 3.72–3.69 (m, 2H), 3.43–3.26 (m, 4H); 13C NMR (75.5 MHz, CDCl3) δ 135.4, 135.3, 128.8, 128.7, 128.1, 70.2, 70.1, 69.9, 69.8, 69.7, 68.54, 68.46, 53.3; 31P NMR (121.5 MHz, CDCl3) δ 0.928; HRMS [M – N2 + H]+ calcd: 380.12630, found: 380.12495.
(2S,3S)-1-azido-4-(bis(benzyloxy)phosphoryloxy)butane-2,3-diyl distearate (2)
Diol 9 (482 mg, 1.18 mmol), stearic acid (1.01 g, 3.55 mmol), dicyclohexylcarbodiimide (732 mg, 3.55 mmol) and 4-dimethylaminopyridine (144 mg, 1.18 mmol) were dissolved in dichloromethane (12 mL) and stirred at room temperature overnight. The reaction was filtered using ethyl acetate and concentrated under reduced pressure. Column chromatography with silica gel and a gradient solvent system of 10–30% ethyl acetate to hexanes afforded 2 as a white solid (820 mg, 74%).
[α]D296K – 4.26 (c = 1.315, CHCl3); 1H NMR (300 MHz, CDCl3) δ 7.35–7.26 (m, 10H), 5.20 (bs, 2H), 5.04–5.01 (m, 4H), 4.13–3.90 (m, 2H), 3.45–3.28 (m, 2H), 2.35–2.24 (m, 4H), 1.65–1.58 (m, 4H), 1.26 (bs, 56H), 0.88 (t, J = 6.5 Hz, 6H); 13C NMR (75.5 MHz, CDCl3) δ 172.7, 172.6, 135.6, 135.5, 128.7, 128.6, 128.03, 127.99, 70.0, 69.9, 69.7, 69.6, 69.5, 65.1, 65.0, 50.6, 34.0, 31.9, 29.72, 29.68, 29.5, 29.4, 29.3, 29.1, 24.81, 24.78, 22.7, 14.1; 31P NMR (121.5 MHz, CDCl3) δ 0.038; MALDI–HRMS [M + Na]+ calcd: 962.6358, found: 962.6332.
Rhodamine-Alkyne Conjugate (10d)
Lissamine Rhodamine B sulfonyl chloride (141 mg, 0.244 mmol) was dissolved in DMF (10 mL). Propargyl amine (17 μL, 0.244 mmol) and triethyl amine (170 µL, 1.22 mmol) were added with stirring. Stirring was continued in the dark at room temperature overnight at which time the solvent was removed under reduced pressure. Column chromatography with silica gel and a gradient solvent system of 2–7% methanol to dichloromethane afforded 10d as a purple solid (55 mg, 38%).
1H NMR (300 MHz, CDCl3) δ 8.74 (m, 1H), 8.13–8.10 (m, 1H), 7.37 (d, J = 8.1 Hz, 1H), 7.15 (d, J = 9.3 Hz, 2H), 6.93–6.89 (m, 2H), 6.82–6.81 (m, 2H), 3.93 (d, J = 2.4 Hz, 2H), 3.64 (q, J = 7.2 Hz, 8H), 2.44 (t, J = 2.5 Hz, 1H), 1.33 (t, J = 7.2 Hz, 12H); 13C NMR (62.9 MHz, CDCl3) δ 158.3, 157.4, 156.0, 146.6, 142.7, 134.4, 133.0, 130.7, 128.6, 127.6, 114.5, 114.1, 96.2, 78.6, 73.5, 46.2, 32.8, 12.7; MALDI–HRMS [M + Na]+ calcd: 618.1703, found: 618.1699
Benzophenone-Lys(Boc)-OMe (14)
Cbz-Lys(Boc)-OMe (13, 535 mg, 1.36 mmol) was dissolved in methanol (15 mL). Palladium (II) hydroxide (10% wt) was added with stirring. A hydrogen atmosphere was established and stirring was continued overnight. The reaction mixture was filtered through celite and washed with methanol and the solvent was removed under reduced pressure. The residue was dissolved in DMF (10 mL). With stirring 4-benzoylbenzoic acid (308 mg, 1.36 mmol), 4-dimethylaminopyridine (199 mg, 1.63 mmol), and N-methylmorpholine (523 µL, 4.76 mmol) were added. After 5 min EDCI·HCl (313 mg, 1.63 mmol) was added and stirring was continued 7 h. The solvent was removed under reduced pressure and column chromatography with silica gel and a solvent system of 50% ethyl acetate to hexanes afforded 14 as a white foam (561 mg, 88%).
[α]D296K + 8.89(c = 5.544, CHCl3); 1H NMR (300 MHz, CDCl3) δ 7.96 ( d, J = 8.4 Hz, 2H), 7.84–7.77 (m, 4H), 7.62 (t, J = 7.5 Hz, 1H), 7.52–7.47 (m, 2H), 7.26 (d, J = 7.5 Hz, 1H), 4.81–4.77 (m, 2H), 3.78 ( s, 3H), 3.13–3.11 (m, 2H), 2.00–1.40 (m, 15H); 13C NMR (100.6 MHz, CDCl3) δ 195.9, 172.9, 166.5, 156.2, 140.3, 137.1, 137.0, 132.9, 130. 23, 130.20, 128.4, 127.2, 79.1, 52.7, 52.5, 40.0, 31.9, 29.7, 28.4, 22.6; HRMS [M - Boc + 2H]+ calcd: 369.18143, found: 369.18007
Benzophenone-Lys(Boc)-Alkyne (10f)
Compound 14 (561 mg, 1.20 mmol) was dissolved in methanol (5 mL). With stirring was added 2 N NaOH (5 mL). Stirring was continued 20 min and TLC analysis showed no starting material. The solution was neutralized with Dowex®50WX8–200 H+ resin to pH ~4. The solution was filtered and the filtrate concentrated under reduced pressure. The residue was washed with toluene, concentrated, and dried under high vacuum 5 h. The residue was then dissolved in dichloromethane (15 mL) and with stirring propargyl amine (99 µL, 1.44 mmol), 4-dimethylaminopyridine (146 mg, 1.20 mmol), EDCI∙HCl (276 mg, 1.44 mmol) and N-methylmorpholine (395 µL, 3.59 mmol) were added. Stirring was continued overnight at room temperature at which time the solvent was removed under reduced pressure. Column chromatography with silica gel and a gradient solvent system of 50–80% ethyl acetate to hexanes afforded 10f as a white solid (550 mg, 93%, 2 steps).
[α]D296K + 1.07(c = 0.935, CHCl3); 1H NMR (300 MHz, CDCl3) δ 7.96–7.92 (m, 2H), 7.81–7.74 (m, 4H), 7.61–7.44 (m, 5H), 4.82–4.75 (m, 2H), 4.12–3.96 (m, 2H), 3.08–3.07 (m, 2H), 2.19 (t, J = 2 Hz, 1H), 2.00–1.77 (m, 2H), 1.59–1.38 (m, 12H); 13C NMR (75.5 MHz, CDCl3) δ 195.9, 171.8, 166.8, 156.2, 140.3, 136.9, 136.8, 132.9, 130.1, 130.0, 128.5, 127.3, 79.3, 79.1, 71.7, 53.5, 40.1, 32.3, 29.6, 29.2, 28.4, 22.7; HRMS [M - Boc + 2H]+ calcd: 392.19742, found: 392.19514
General Procedure for Click Chemistry Derivatization to Produce Phosphotriesters 11a–e
Compound 2 and derivatized alkynes (10a–e) were dissolved in THF (1.5–2 mL). Copper sulfate pentahydrate and sodium ascorbate were added along with H2O (0.5 mL). The solution was stirred at room temperature overnight at which point the solvent was removed under reduced pressure. Column chromatography with silica gel afforded compounds 11a–e. Due to the amphipathic nature of the phosphotriesters, the integration values in the proton NMR spectra sometimes varied slightly from the expected values.
Naphthyl Phosphotriester (11a)
Alkyne 10a (33.7 mg, 0.151 mmol), 2 (141.6 mg, 0.151 mmol), copper sulfate pentahydrate (113 mg, 0.453 mmol), and sodium ascorbate (179 mg, 0.906 mmol) were utilized. Column chromatography with a gradient solvent system of 70–100% ethyl acetate to hexanes afforded 11a as a white solid (151 mg, 86%).
[α]D296K – 4.41 (c = 3.79, CHCl3); 1H NMR (300 MHz, CDCl3) δ 7.80–7.75 (m, 3H), 7.69 (s, 1H), 7.48–7.43 (m, 3H), 7.36–7.33 (m, 11H), 6.52 (t, J = 5.4 Hz, 1H), 5.44–5.39 (m, 1H), 5.03–4.97 (m, 5H(1 + 4 H)), 4.43–4.41 (m, 4H), 4.18–4.06 (m, 2H), 3.71 (s, 2H), 2.29–2.18 (m, 4H), 1.56–1.48 (m, 4H), 1.25 (bs, 56H), 0.88 (t, J = 6 Hz, 6H); 13C NMR (75.5 MHz, CDCl3) δ 172.6, 172.2, 171.1, 144.9, 135.5, 135.4, 133.5, 132.4, 132.2, 128.7, 128.6, 128.1, 128.0, 127.96, 127.3, 126.2, 125.9, 123.5, 69.65, 69.57, 69.1, 64.82, 64.81, 49.9, 49.8, 43.6, 34.9, 33.9, 33.8, 31.9, 29.71, 29.67, 29.5, 29.4, 29.3, 29.12, 29.07, 24.8, 24.7, 22.7, 14.1; 31P NMR (121.5 MHz, CDCl3) δ -0.001; MALDI–HRMS [M + Na]+ calcd: 1185.7355, found: 1185.7310.
Dansyl Phosphotriester (11b)
Alkyne 10b (33.4 mg, 0.116 mmol), 2 (109 mg, 0.116 mmol), copper sulfate pentahydrate (87 mg, 0.348 mmol), and sodium ascorbate (138 mg, 0.696 mmol) were utilized. Column chromatography with a gradient solvent system of 50–80% ethyl acetate to hexanes afforded 11b as a green glass (121 mg, 85%).
[α]D296K – 4.40 (c = 1.796, CHCl3); 1H NMR (300 MHz, CDCl3) δ 8.55 (d, J = 8.7 Hz, 1H), 8.29–8.25 (m, 2H), 7.53 (q, J = 7.9 Hz, 2H), 7.34 (bs, 11H), 7.18 (d, J = 7.2 Hz, 1H), 5.52 (bs, 1H), 5.40–5.35 (m, 1H), 5.08–4.97 (m, 5H(1 + 4H)), 4.39–4.37 (m, 2H), 4.17–4.05 (m, 4H), 2.89 (s, 6H), 2.30–2.21 (m, 4H), 1.56–1.50 (m, 4H), 1.25 (bs, 56H), 0.88 (t, J = 6.6 Hz, 6H); 13C NMR (75.5 MHz, CDCl3) δ 172.6, 172.3, 151.9, 144.4, 135.45, 135.46, 134.6, 130.5, 129.9, 129.6, 129.5, 128.7, 128.6, 128.5, 128.03, 127.99, 123.4, 123.1, 118.8, 115.3, 69.7, 69.6, 69.1, 64.92, 64.86, 49.8, 45.4, 38.7, 33.9, 33.8, 31.9, 29.72, 29.67, 29.5, 29.4, 29.29, 29.27, 29.13, 29.07, 24.8, 24.7, 22.7, 14.1; 31P NMR (121.5 MHz, CDCl3) δ -0.058; MALDI–HRMS [M + Na]+ calcd: 1250.7290, found: 1250.7317.
Coumarin Phosphotriester (11c)
Alkyne 10c (36.1 mg, 0.159 mmol), 2 (149.3 mg, 0.159 mmol) copper sulfate pentahydrate (119 mg, 0.477 mmol), and sodium ascorbate (189 mg, 0.954 mmol) were employed. Column chromatography with a gradient solvent system of 70–100% ethyl acetate to hexanes afforded 11c as a white solid (153 mg, 83%).
[α]D296K – 6.96 (c = 6.132, CHCl3); 1H NMR (300 MHz, CDCl3) δ 9.28–9.25 (m, 1H) 8.90 (s, 1H), 7.69–7.64 (m, 2H), 7.58 (s, 1H), 7.41–7.34 (m, 12H), 5.49–5.44 (m, 1H), 5.09–4.98 (m, 5H(1+4H)), 4.73–4.71 (m, 2H), 4.49–4.47 (m, 2H), 4.19–4.09 (m, 2H), 2.30–2.22 (m, 4H), 1.57–1.42 (m, 4H), 1.25 (bs, 56H), 0.88 (t, J = 6.5 Hz, 6H); 13C NMR (100.6 MHz, CDCl3) δ 172.5, 172.2, 161.7, 161.1, 154.5, 148.4, 144.9, 135.6, 135.5, 134.2, 129.8, 128.7, 129.0, 125.3, 123.3, 115.5, 118.3, 116.7, 69.7, 69.2, 64.9, 49.9, 35.5, 34.0, 33.8, 32.9, 29.7, 29.5, 29.4, 29.3, 29.2, 24.8, 24.7, 22.7, 14.1; 31P NMR (121.5 MHz, CDCl3) δ 0.027; MALDI–HRMS [M + Na]+ calcd: 1189.6940, found: 1189.6920.
Rhodamine Phosphotriester (11d)
Alkyne 10d (21.4 mg, 0.036 mmol), 2 (34 mg, 0.036 mmol) copper sulfate pentahydrate (27 mg, 0.108 mmol), and sodium ascorbate (43 mg, 0.215 mmol) were used. Column chromatography with 10% methanol / chloroform as eluant afforded 11d as a pink solid (44 mg, 80%).
No optical rotation was obtained due to the high propensity of the compound to stain; 1H NMR (300 MHz, CDCl3) δ 8.52 (s, 1H), 7.93–7.91 (m, 2H), 7.30 (bs, 12H), 7.10 (d, J = 7.5 Hz, 1H), 6.90 (d, J = 8.7 Hz, 2H), 6.63–6.62 (m, 3H), 5.47 (bs, 1H), 5.01–4.97 (m, 5H(1 + 4H)), 4.73–4.69 (m, 1H), 4.58–4.45 (m. 3H), 4.16 (s, 2H), 3.65–3.52 (m, 8H), 2.21–2.18 (m, 4H), 1.47–1.45 (m, 4H), 1.31–1.20 (m, 68H), 0.88 (t, J = 6.6 Hz, 6H); 13C NMR (100.6 MHz, CDCl3) δ 172.6, 172.3, 159.0, 157.8, 155.7, 155.5, 147.4, 142.3, 135.83, 135.76, 133.73, 133.67, 133.3, 129.6, 128.6, 128.5, 128.4, 128.0, 127.4, 127.2, 125.1, 114.3, 113.7, 113.6, 95.6, 70.0, 69.9, 69.63, 69.57, 69.5, 69.4, 68.9, 65.2, 50.1, 45.8, 38.8, 34.1, 33.9, 31.9, 29.7, 29.5, 29.4, 29.1, 24.8, 24.7, 22.7, 14.1, 12.6; 31P NMR (121.5 MHz, CDCl3) δ -0.247; MALDI–HRMS [M + Na]+ calcd: 1557.8169, found: 1557.8251.
Benzophenone Phosphotriester (11e)
Alkyne 10e (38 mg, 0.144 mmol), 2 (123 mg, 0.131 mmol), copper sulfate pentahydrate (98 mg, 0.392 mmol), and sodium ascorbate (155 mg, 0.785 mmol) were used. Column chromatography with a gradient solvent system of 70–100% ethyl acetate to hexanes afforded 11e as a white solid (135 mg, 86%).
[α]D296K – 5.27 (c = 2.544, CHCl3); 1H NMR (300 MHz, CDCl3) δ 7.94 (d, J = 7.8 Hz, 2H), 7.83–7.76 (m, 4H), 7.64–7.58 (m, 2H), 7.48 (t, J = 7.7 Hz, 3H), 7.33 (bs, 10H), 5.49–5.43 (m, 1H), 5.11–4.96 (m, 5H(1 + 4H)), 4.78–4.71 (m, 2H), 4.55–4.49 (m, 2H), 4.19–4.09 (m, 2H), 2.30–2.22 (m. 4H), 1.56–1.49 (m, 4H), 1.25 (bs, 56H), 0.88 (t, J = 6.5 Hz, 6H); 13C NMR (100.6 MHz, CDCl3) δ 195.8, 172.6, 172.2, 166.5, 140.1, 137.2, 137.1, 135.54, 135.47, 132.8, 130.0, 128.6, 128.4, 128.0, 127.97, 127.1, 123.6, 69.7, 69.6, 69.2, 64.9, 64.8, 50.0, 35.5, 34.0, 33.8, 31.9, 29.7, 29.5, 29.4, 29.3, 29.14, 29.10, 24.8, 24.7, 22.7, 14.1; 31P NMR (121.5 MHz, CDCl3) δ -0.058; MALDI–HRMS [M + H]+ calcd: 1203.7485, found: 1203.7488.
Bifunctional benzophenone/azido Phosphotriester (11f)
Alkyne 10f (84 mg, 0.171 mmol) was dissolved in dichloromethane (2 mL) and with stirring was added trifluoroacetic acid (2 mL). Stirring was continued for 2 h and the solvent was removed under reduced pressure and placed under high vacuum for 2 h. The residue was dissolved in THF (1.5 mL) and 2 (161 mg, 0.171 mmol) was added along with copper sulfate pentahydrate (43 mg, 0.171 mmol), sodium ascorbate (68 mg, 0.342 mmol), and H2O (0.5 mL). Stirring was continued overnight at room temperature at which point the solvent was removed under reduced pressure and placed under high vacuum 2 h. This residue was dissolved in anhydrous DMF (3 mL) and 6-azidohexanoic acid71 (32 mg, 0.205 mmol) was added. 4-Dimethylaminopyridine (21 mg, 0.171 mmol), EDCI∙HCl (72 mg, 0.376 mmol), and N-methylmorpholine (94 µL, 0.855 mmol) were added and stirring was continued overnight at room temperature at which point the solvent was removed under reduced pressure. Column chromatography with silica gel and a gradient solvent system of 30–50% acetone to chloroform afforded 11f as a white solid (110 mg, 44%, 3 steps).
[α]D296K – 4.91 (c = 4.36, CHCl3); 1H NMR (300 MHz, CDCl3) δ 7.98 (d, J = 8.4 Hz, 2H), 7.80 (t J = 8.4 Hz, 4H), 7.64–7.47 (m, 6H), 7.34 (bs, 10H), 6.16–6.12 (m, 1H), 5.47–5.42 (m, 1H), 5.07–4.96 (m, 5H(1 + 4H)), 4.68–4.66 (m, 1H), 4.52–4.52 (m, 4H), 4.13–4.07 (m, 2H), 3.26–3.19 (m, 4H), 2.30–2.23 (m, 4H), 2.12 (t, J = 7.5 Hz, 2H), 1.96–1.84 (m, 2H), 1.63–1.24 (m, 70H), 0.88 (t J = 6.5 Hz, 6H); 13C NMR (100.6 MHz, CDCl3) δ 195.8, 173.2, 172.6, 173.3, 171.8, 166.7, 144.8, 140.3, 137.0, 135.5, 135.4, 132.9, 130.1, 130.0, 128.72, 128.67, 128.4, 128.0, 127.96, 127.3, 123.3, 69.72, 69.67, 69.2, 64.9, 53.6, 51.2, 49.9, 38.6, 36.4, 35.1, 34.0, 33.9, 31.9, 29.7, 29.6, 29.4, 29.3, 29.14, 29.06, 28.6, 26.4, 25.2, 24.80, 24.76, 22.7, 22.5, 14.1; 31P NMR (121.5 MHz, CDCl3) δ -0.254; MALDI–HRMS [M + Na]+ calcd: 1492.8999, found: 1492.9028.
General Procedure for Deprotection of Phosphotriesters to Access PA Analogues 1b / 12a–f
Phosphotriesters 2 / 11a–f were dissolved in dichloromethane at 0 °C under argon. Bromotrimethylsilane was added and stirring was continued at room temperature for 1 h. Solvent was removed under reduced pressure and the residue was placed on a high vacuum line to remove any residual solvents. Methanol (2 mL) was added and the solution was stirred at room temperature for 1 h. The solid that formed was collected via filtration. The solid was dissolved in methanol–chloroform (1:4) and extracted with 8 mM ammonium acetate. The organic layer was concentrated under reduced pressure to afford 1b / 12a–f.
Azido PA (1b)
Azido phosphotriester 2 (65 mg, 0.069 mmol), dichloromethane (2.45 mL), and bromotrimethylsilane (0.55 mL) afforded 1b as a white solid (30 mg, 56%). 1H NMR (300 MHz, CDCl3 : CD3OD (1:1)) δ 5.29 (bs, 2H), 4.09 (bs, 2H), 3.61–3.52 (m, 2H), 2.38–2.37 (m, 4H), 1.64 (bs, 4H), 1.27 (bs, 56H), 0.89 (t, J = 6.3 Hz, 6H); 31P NMR showed no resonance in any solvent tried; MALDI–HRMS [M - NH4 + 2Na]+ calcd: 804.5238, found: 804.5143.
Naphthyl PA (12a)
Naphthyl phosphotriester 11a (67 mg, 0.058 mmol), dichloromethane (1.64 mL), and bromotrimethylsilane (0.36 mL) afforded 12a as a brownish solid (40 mg, 70%).
31P NMR (121.5 MHz, CDCl3) δ 1.44; MALDI–HRMS [M - NH4 + 2Na]+ calcd: 1027.6235, found: 1027.6273.
Dansyl PA (12b)
Dansyl phosphotriester 11b (80 mg, 0.065 mmol), dichloromethane (1.64 mL), and bromotrimethylsilane (0.36 mL) afforded 12b as a green solid (74 mg, quantative yield).
31P NMR (121.5 MHz, CDCl3) δ 1.09; MALDI–HRMS [M - NH4 + 2Na]+ calcd: 1092.6171, found: 1092.6128.
Coumarin PA (12c)
Coumarin phosphotriester 11c (60 mg, 0.051 mmol), dichloromethane (1.64 mL), and bromotrimethylsilane (0.36 mL) afforded 12c as a white solid (40 mg, 78%).
31P NMR (121.5 MHz, CDCl3) δ 1.39; MALDI–HRMS [M - NH4 + 2Na]+ calcd: 1031.5821, found: 1031.5857.
Rhodamine PA (12d)
Rhodamine phosphotriester 11d (44mg, 0.029 mmol), dichloromethane (1.64 mL), and bromotrimethylsilane (0.36 mL) Column chromatography with silica gel and a solvent system of 7 chloroform : 2.5 methanol : 0.5 water was necessary to purify the phosphate. The resulting solid was dissolved in methanol–chloroform (1:4) and extracted with 8 mM ammonium acetate. The organic layer was concentrated under reduced pressure to afford 12d as a purple solid (34 mg, 85%).
31P NMR no resonance could be detected in any solvent tried; MALDI–HRMS [M - NH4 + 2Na]+ calcd: 1399.7049, found: 1399.6963.
Benzophenone PA (12e)
Benzophenone phosphotriester 11e (41.5 mg, 0.035 mmol), dichloromethane (1.64 mL), and bromotrimethylsilane (0.36 mL) afforded 12e as a pinkish white solid (26 mg, 73%).
31P NMR (121.5 MHz, CDCl3) δ 1.25; MALDI–HRMS [M - NH4 + 2Na]+ calcd: 1067.6185, found: 1067.6116.
Bifunctional Benzophenone/Azido PA (12f)
Bifunctional benzophenone/azido phosphotriester 11f (52 mg, 0.035 mmol), dichloromethane (1.64 mL), and bromotrimethylsilane (0.36 mL) afforded 12f as a white solid (38 mg, 83%).
31P NMR (121.5 MHz, CDCl3) δ 1.12; MALDI–HRMS [M - NH4 + 2Na]+ calcd: 1334.7880, found: 1334.7890.
PA Binding Analysis with Surface Plasmon Resonance
All surface plasmon resonance (SPR) binding measurements were performed at room temprature (25°C). PA lipid stocks were dissolved in a chloroform:methanol (80:20) mixture from which liposomes were prepared as POPC:POPE:PA (40:40:20) for different PA compounds. Aliquots of lipid mixtures to prepare a final concentration of 0.5 mg/ml of vesicles were dried under nitrogen and rehydrated with 500 µl binding buffer (25 mM Tris HCl pH 7.4, 150 mM NaCl, 10 µM CaCl2 and 1 mM DTT). Lipid solutions were vortexed for 15 min and sonicated for 15 min. Lipid vesicles were then passed 19 times through a 100 nm polycarbonate membrane in an Avanti Mini-Extruder at room temperature. The liposome coating of the L1 sensor chip (GE Healthcare) has been described in detail previously.74,75 Briefly, the sensor chip surface was washed and then coated by injecting 85 µl of vesicles containing various analogs of PA at a 5 µl/min flow rate to yield a response of 5,000 resonance units (RU). Each lipid layer was stabilized by injecting 10 µl of 50 mM NaOH (the regeneration solution) three times at 100 µl/min flow rate. Typically, no decrease in lipid signal was seen after the first injection. Various concentrations of PKCα-C2 (within a 10-fold range of Kd) were injected to study the equilibrium binding and viability of synthetic analogs of PA. PKCα-C2 was expressed and purified as previously described.76 For Equilibrium SPR measurements, the flow rate was set at 10 µl/min to allow sufficient time for the association phase, which in turn allows RU values to reach saturating response values (Req). Following binding of each respective injection of PKCα-C2, the lipid layer was regenerated to baseline with two 30 µl pulses of 50 mM NaOH injected at a flow rate of 30 µl/min. Following collection of three data sets for each PA analogue, Req values were plotted versus respective protein concentrations (C), and the Kd value was calculated by a non-linear least squares analysis of the binding isotherm using an equation, Req = Rmax/(1 + Kd/C),75 where Rmax is a theoretical maximum Req value. Validation of Kd values determined from equilibrium binding response values was performed by analyzing sensorgrams with BIAevaluation software to determine rate constants of association (ka) and (kd) as described previously,78 assuming a 1:1 protein-membrane surface binding, because all kinetic sensorgrams agreed with this model. Furthermore, Kd values determined by kinetic fits were similar (±15%) to Kd values obtained through equilibrium binding analysis.
Supplementary Material
Acknowledgments
MDB acknowledge funding from the University of Tennessee at Knoxville and RVS acknowledges a grant from the American Heart Association (SDG 0735350N).
References
- 1.Sakane F, Imai S, Kai M, Yasuda S, Kanoh H. Biochim. Biophys. Acta. 2007;1771:793–806. doi: 10.1016/j.bbalip.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Stace CL, Ktistakis NT. Biochim. Biophys. Acta. 2006;1761:913–926. doi: 10.1016/j.bbalip.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Testerink C, Munnik T. Trends Plant Sci. 2005;10:368–375. doi: 10.1016/j.tplants.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Colon-Gonzalez F, Kazanietz MG. Biochim. Biophys. Acta. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Cho WH, Stahelin RV. Ann. Rev. Biophys. Biomol. Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 6.Di Paolo G, De Camilli P. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 7.Lemmon MA. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 8.Lemmon MA. Nature Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 9.Misra S, Miller GJ, Hurley JH. Cell. 2001;107:559–562. doi: 10.1016/s0092-8674(01)00594-3. [DOI] [PubMed] [Google Scholar]
- 10.Overduin M, Cheever ML. Mol. Intervent. 2001;1:150–159. [PubMed] [Google Scholar]
- 11.Prestwich GD. Chem. Biol. 2004;11:619–637. doi: 10.1016/j.chembiol.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 12.Sprong H, van der Sluijs P, van Meer G. Nat. Rev. Mol. Cell Biol. 2001;2:698–698. doi: 10.1038/35080071. [DOI] [PubMed] [Google Scholar]
- 13.Hurley JH. Biochim. Biophys. Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurley JH, Misra S. Annu. Rev. Biophys. Biomolec. Struct. 2000;29:49–79. doi: 10.1146/annurev.biophys.29.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta S, Ramjaun AR, Haiko P, Wang J, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 16.Wittinghofer A, Waldmann H. Angew. Chem., Int. Edit. 2000;39:4193–4214. doi: 10.1002/1521-3773(20001201)39:23<4192::AID-ANIE4192>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 17.Fang YM, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 18.Jenkins GH, Fisette PL, Anderson RA. J. Biol. Chem. 1994;269:11547–11554. [PubMed] [Google Scholar]
- 19.Moritz A, Degraan PNE, Gispen WH, Wirtz KWA. J. Biol. Chem. 1992;267:7207–7210. [PubMed] [Google Scholar]
- 20.Frank C, Keilhack H, Opitz F, Zschornig O, Bohmer FD. Biochemistry. 1999;38:11993–12002. doi: 10.1021/bi982586w. [DOI] [PubMed] [Google Scholar]
- 21.Jones JA, Hannun YA. J. Biol. Chem. 2002;277:15530–15538. doi: 10.1074/jbc.M111555200. [DOI] [PubMed] [Google Scholar]
- 22.Jones JA, Rawles R, Hannun YA. Biochemistry. 2005;44:13235–13245. doi: 10.1021/bi0505159. [DOI] [PubMed] [Google Scholar]
- 23.Baillie GS, Huston E, Scotland G, Hodgkin M, Gall I, Peden AH, MacKenzie C, Houslay ES, Currie R, Pettitt TR, Walmsley AR, Wakelam MJO, Warwicker J, Houslay MD. J. Biol. Chem. 2002;277:28298–28309. doi: 10.1074/jbc.M108353200. [DOI] [PubMed] [Google Scholar]
- 24.Grange M, Sette C, Cuomo M, Conti M, Lagarde M, Prigent AF, Nemoz G. J. Biol. Chem. 2000;275:33379–33387. doi: 10.1074/jbc.M006329200. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh S, Strum JC, Sciorra VA, Daniel L, Bell RM. J. Biol. Chem. 1996;271:8472–8480. doi: 10.1074/jbc.271.14.8472. [DOI] [PubMed] [Google Scholar]
- 26.Andresen BT, Rizzo MA, Shome K, Romero G. FEBS Lett. 2002;531:65–68. doi: 10.1016/s0014-5793(02)03483-x. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh S, Moore S, Bell RM, Dush M. J. Biol. Chem. 2003;278:45690–45696. doi: 10.1074/jbc.M302933200. [DOI] [PubMed] [Google Scholar]
- 28.Hekman M, Hamm H, Villar AV, Bader B, Kuhlmann J, Nickel J, Rapp UR. J. Biol. Chem. 2002;277:24090–24102. doi: 10.1074/jbc.M200576200. [DOI] [PubMed] [Google Scholar]
- 29.Improta-Brears T, Ghosh S, Bell RM. Mol. Cell. Biochem. 1999;198:171–178. doi: 10.1023/a:1006981411691. [DOI] [PubMed] [Google Scholar]
- 30.Mott HR, Carpenter JW, Zhong S, Ghosh S, Bell RM, Campbell SL. Proc. Natl. Acad. Sci. U.S.A. 1996;93:8312–8317. doi: 10.1073/pnas.93.16.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizzo MA, Shome K, Vasudevan C, Stolz DB, Sung TC, Frohman MA, Watkins SC, Romero G. J. Biol. Chem. 1999;274:1131–1139. doi: 10.1074/jbc.274.2.1131. [DOI] [PubMed] [Google Scholar]
- 32.Rizzo MA, Shome K, Watkins SC, Romero G. J. Biol. Chem. 2000;275:23911–23918. doi: 10.1074/jbc.M001553200. [DOI] [PubMed] [Google Scholar]
- 33.Ochoa WF, Corbalan-Garcia S, Eritja R, Rodriguez-Alfaro JA, Gomez-Fernandez JC, Fita I, Verdaguer N. J. Mol. Biol. 2002;320:277–291. doi: 10.1016/S0022-2836(02)00464-3. [DOI] [PubMed] [Google Scholar]
- 34.Ochoa WF, Garcia-Garcia J, Corbalan-Garcia IFS, Verdaguer N, Gomez-Fernandez JC. J. Mol. Biol. 2001;311:837–849. doi: 10.1006/jmbi.2001.4910. [DOI] [PubMed] [Google Scholar]
- 35.Carrasco S, Merida I. Trends Biochem. Sci. 2007;32:27–36. doi: 10.1016/j.tibs.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Gomez-Fernandez JC, Corbalan-Garcia S. Chem. Phys. Lipids. 2007;148:1–25. doi: 10.1016/j.chemphyslip.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Spitaler M, Cantrell DA. Nat. Immunol. 2004;5:785–790. doi: 10.1038/ni1097. [DOI] [PubMed] [Google Scholar]
- 38.Dorn GW, Mochly-Rosen D. Annu. Rev. Physiol. 2002;64:407–429. doi: 10.1146/annurev.physiol.64.081501.155903. [DOI] [PubMed] [Google Scholar]
- 39.Griner EM, Kazanietz MG. Nat. Rev. Cancer. 2007;7:281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 40.Gutcher I, Webb PR, Anderson NG. Cell. Mol. Life Sci. 2003;60:1061–1070. doi: 10.1007/s00018-003-2281-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mackay HJ, Twelves CJ. Nat. Rev. Cancer. 2007;7:554–562. doi: 10.1038/nrc2168. [DOI] [PubMed] [Google Scholar]
- 42.Schultz A, Ling M, Larsson C. J. Biol. Chem. 2004;279:31750–31760. doi: 10.1074/jbc.M313017200. [DOI] [PubMed] [Google Scholar]
- 43.Kooijman EE, Tieleman DP, Testerink C, Munnik T, Rijkers DTS, Burger KNJ, de Kruijff B. J. Biol. Chem. 2007;282:11356–11364. doi: 10.1074/jbc.M609737200. [DOI] [PubMed] [Google Scholar]
- 44.Kooijman EE, Carter KM, van Laar EG, Chupin V, Burger KNJ, de Kruijff B. Biochemistry. 2005;44:17007–17015. doi: 10.1021/bi0518794. [DOI] [PubMed] [Google Scholar]
- 45.Hanshaw RG, Stahelin RV, Smith BD. Chem. Eur. J. 2008;14:1690–1697. doi: 10.1002/chem.200701589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Picq M, Huang Y, Lagarde M, Doutheau A, Nemoz G. J. Med. Chem. 2002;45:1678–1685. doi: 10.1021/jm011032f. [DOI] [PubMed] [Google Scholar]
- 47.Lim ZY, Thuring JW, Holmes AB, Manifava M, Ktistakis NT. J. Chem. Soc., Perkin Trans. 1. 2002:1067–1075. [Google Scholar]
- 48.Manifava M, Thuring J, Lim ZY, Packman L, Holmes AB, Ktistakis NT. J. Biol. Chem. 2001;276:8987–8994. doi: 10.1074/jbc.M010308200. [DOI] [PubMed] [Google Scholar]
- 49.Xu Y, Fang YM, Chen J, Prestwich GD. Bioorg. Med. Chem. Lett. 2004;14:1461–1464. doi: 10.1016/j.bmcl.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 50.Goedhart J, Gadella TWJ. Biochemistry. 2004;43:4263–4271. doi: 10.1021/bi0351460. [DOI] [PubMed] [Google Scholar]
- 51.Neef AB, Schultz C. Angew. Chem., Int. Ed. 2009;48:1498–1500. doi: 10.1002/anie.200805507. [DOI] [PubMed] [Google Scholar]
- 52.Rzepecki PW, Prestwich GD. J. Org. Chem. 2002;67:5454–5460. doi: 10.1021/jo011185a. [DOI] [PubMed] [Google Scholar]
- 53.Smith MD, Gong D, Sudhahar CG, Reno JC, Stahelin RV, Best MD. Bioconj. Chem. 2008;19:1855–1863. doi: 10.1021/bc8001002. [DOI] [PubMed] [Google Scholar]
- 54.Kolb HC, Finn MG, Sharpless KB. Angew. Chem.-Int. Edit. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 55.Lewis WG, Green LG, Grynszpan F, Radic Z, Carlier PR, Taylor P, Finn MG, Sharpless KB. Angew. Chem., Int. Edit. 2002;41:1053–1057. doi: 10.1002/1521-3773(20020315)41:6<1053::aid-anie1053>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 56.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem., Int. Edit. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 57.O'Neil EJ, DiVittorio KM, Smith BD. Org. Lett. 2007;9:199–202. doi: 10.1021/ol062557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lampkins AJ, O'Neil EJ, Smith BD. J. Org. Chem. 2008;73:6053–6058. doi: 10.1021/jo8011336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahn T, Oh DB, Kim H, Park C. J. Biol. Chem. 2002;277:26157–26162. doi: 10.1074/jbc.M203445200. [DOI] [PubMed] [Google Scholar]
- 60.Ahn T, Oh DB, Lee BC, Yun CH. Biochemistry. 2000;39:10147–10153. doi: 10.1021/bi0000622. [DOI] [PubMed] [Google Scholar]
- 61.Kim KH, Ahn T, Yun CH. Biochemistry. 2003;42:15377–15387. doi: 10.1021/bi035280k. [DOI] [PubMed] [Google Scholar]
- 62.Song XD, Nolan J, Swanson BI. J. Am. Chem. Soc. 1998;120:11514–11515. [Google Scholar]
- 63.Song XD, Shi J, Nolan J, Swanson B. Anal. Biochem. 2001;291:133–141. doi: 10.1006/abio.2001.5024. [DOI] [PubMed] [Google Scholar]
- 64.Dorman G, Prestwich GD. Trends Biotechnol. 2000;18:64–77. doi: 10.1016/s0167-7799(99)01402-x. [DOI] [PubMed] [Google Scholar]
- 65.Cravatt BF, Wright AT, Kozarich JW. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 66.Evans MJ, Cravatt BF. Chem. Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 67.Salisbury CM, Cravatt BF. QSAR Comb. Sci. 2007;26:1229–1238. [Google Scholar]
- 68.Ballel L, Alink K, Slijper M, Versluis C, Liskamp RM, Pieters RJ. ChemBioChem. 2005;6:291–295. doi: 10.1002/cbic.200400209. [DOI] [PubMed] [Google Scholar]
- 69.Ballell L, van Scherpenzeel M, Buchalova K, Liskamp RMJ, Pieters RJ. Org. Biomol. Chem. 2006;4:4387–4394. doi: 10.1039/b611050a. [DOI] [PubMed] [Google Scholar]
- 70.Gubbens J, Ruijter E, de Fays LEV, Damen JM, de Kruijff B, Slijper M, Rijkers DTS, Liskamp RMJ, de Kroon AIPM. Chem. Biol. 2009;16:3–14. doi: 10.1016/j.chembiol.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 71.Grandjean C, Boutonnier A, Guerreiro C, Fournier JM, Mulard LA. J. Org. Chem. 2005;70:7123–7132. doi: 10.1021/jo0505472. [DOI] [PubMed] [Google Scholar]
- 72.Garcia-Garcia J, Corbalan-Garcia S, Gomez-Fernandez JC. Biochemistry. 1999;38:9667–75. doi: 10.1021/bi9905765. [DOI] [PubMed] [Google Scholar]
- 73.Wroblewski AE, Glowacka IE. Tetrahedron. 2005;61:11930–11938. [Google Scholar]
- 74.Bittova L, Stahelin RV, Cho W. J. Biol. Chem. 2001;276:4218–4226. doi: 10.1074/jbc.M008491200. [DOI] [PubMed] [Google Scholar]
- 75.Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho WH. J. Biol. Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- 76.Stahelin RV, Rafter JD, Das S, Cho W. J. Biol. Chem. 2003;278:12452–12460. doi: 10.1074/jbc.M212864200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.