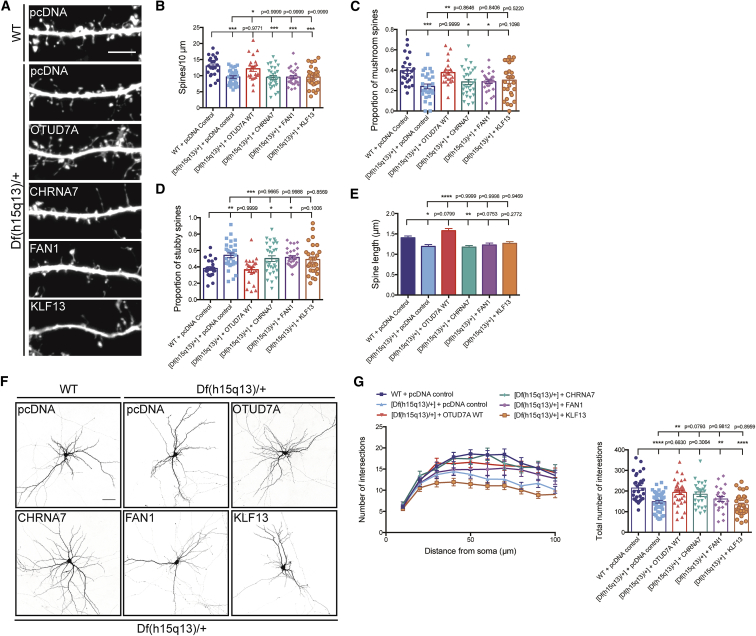
Figure 6.
OTUD7A Is the Predominant Gene in the 15q13.3 Microdeletion Regulating Dendritic Spine Development
(A) Dendritic spines from DIV14 WT and Df(h15q13)/+ cortical neurons co-expressing Venus and the indicated construct (scale bar = 5 μm).
(B–E) Expression of CHRNA7, FAN1, or KLF13 in Df(h15q13)/+ neurons did not rescue the defects in certain phenotypes.
(B) Dendritic spine density. WT + pcDNA control, n = 23 dendritic segments; [Df(h15q13)/+] + pcDNA control, n = 24 dendritic segments; [Df(h15q13)/+] + OTUD7A WT, n = 22 dendritic segments; [Df(h15q13)/+] + CHRNA7, n = 30 dendritic segments; [Df(h15q13)/+] + FAN1, n = 24 dendritic spines; [Df(h15q13)/+] + KLF13, n = 30 dendritic segments; 3 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, F(5, 157) = 6.907.
(C and D) Proportion of mushroom spines (C) and the proportion of stubby spines (D). WT + pcDNA control, n = 23 dendritic segments; [Df(h15q13)/+] + pcDNA control, n = 24 dendritic segments; [Df(h15q13)/+] + OTUD7A WT, n = 22 dendritic segments; [Df(h15q13)/+] + CHRNA7, n = 30 dendritic segments; [Df(h15q13)/+] + FAN1, n = 24 dendritic spines; [Df(h15q13)/+] + KLF13, n = 30 dendritic segments; 3 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, ∗∗p < 0.01, F(5, 156) = 5.045, F(5, 157) = 5.666.
(E) Spine length. WT + pcDNA control, n = 484 spines; [Df(h15q13)/+] + pcDNA control, n = 379 spines; [Df(h15q13)/+] + OTUD7A WT, n = 444 spines; [Df(h15q13)/+] + CHRNA7, n = 454 spines; [Df(h15q13)/+] + FAN1, n = 374 spines; [Df(h15q13)/+] + KLF13, n = 457 spines; 3 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, F(5, 2728) = 10.64.
(F) DIV14 WT and Df(h15q13)/+ cortical neurons co-expressing Venus and the indicated construct (scale bar = 50 μm).
(G) Expression of OTUD7A WT or CHRNA7 in Df(h15q13)/+ cortical neurons rescues dendritic arborization defects, whereas FAN1 and KLF13 did not. WT + pcDNA, n = 30 neurons, [Df(h15q13)/+] + pcDNA control, n = 40 neurons; [Df(h15q13)/+] + OTUD7A WT, n = 35 neurons; [Df(h15q13)/+] + CHRNA7, n = 26 neurons; [Df(h15q13)/+] + FAN1, n = 23 neurons; [Df(h15q13)/+] + KLF13, n = 33 neurons, 3 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, F(5, 181) = 6.873. Error bars represent SEM.