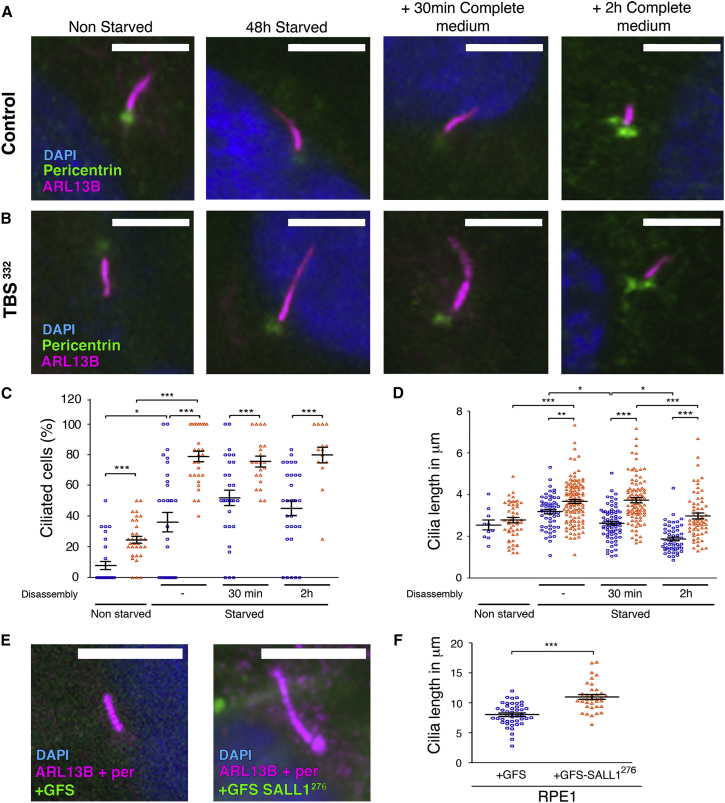
Figure 1.
TBSp.Pro332Hisfs∗10 Cells Show Aberrant Cilia Frequency and Length
(A and B) Micrographs of control HFFs (A) or TBSp.Pro332Hisfs∗10 cells (TBS332) (B) analyzed during cilia assembly and disassembly. Cilia were visualized by ARL13B (purple), basal bodies were visualized by pericentrin (green), and nuclei were visualized by DAPI (blue).
(C and D) Graphical representation of cilia frequency (C) and cilia length (D) measured in control HFFs (blue circles; n = 58 cilia) or TBS332 cells (orange triangles; n = 116 cilia) from three independent experiments. Cells that underwent 48 hr of starvation were compared with non-starved cells. After starvation, cells were supplied with serum for 30 min or 2 hr (n = 21–30 micrographs for all cases).
(E) Micrographs of RPE1 cells infected with lentivirus expressing GFS alone (left) or GFS-SALL1c.826C>T (GFS-SALL1276; right) and labeled with ARL13B (purple) and DAPI (blue).
(F) Graphical representation of cilia length measured in control (blue circles; n = 43 cilia) or GFS-SALL1276 (orange triangles; n = 36 cilia) RPE1 cells infected as in (E).
Graphs represent mean and SEM of three independent experiments. p values were calculated with a two-tailed unpaired Student’s t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars, 5 μm.