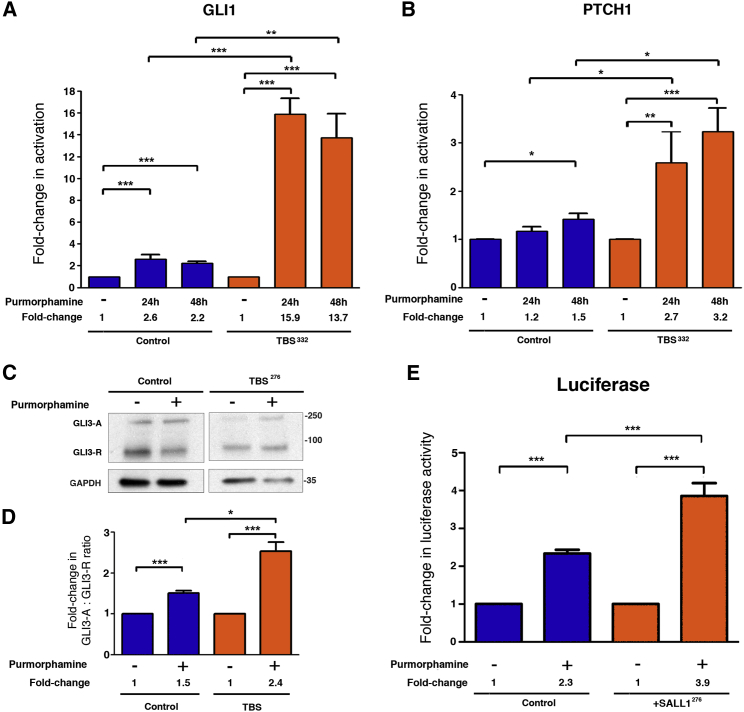
Figure 3.
TBSp.Pro332Hisfs∗10 Fibroblasts Show Aberrant SHH Signaling
(A and B) Graphical representation of the fold change of activation based on the relative expression of GLI1 (A) and PTCH1 (B) obtained by qPCR from control HFFs (n = 4; blue bars) or TBSp.Pro332Hisfs∗10 fibroblasts (TBS332) (n = 4; orange bars) either treated or not (−) with purmorphamine for the indicated times. Numerical values for the fold change are indicated.
(C) Western blot analysis of lysates from control ESCTRL#2 and TBSp.Arg276∗ (TBS276). Samples were probed against GLI3 (A, activator; R, repressor), and GAPDH was used as a loading control. Molecular-weight markers are shown to the right.
(D) Graphical representation is shown for the fold change of the GLI3-A:GLI3-R ratios obtained in (C) for control and TBS fibroblasts. Data from at least three independent experiments using ESCTRL#2 (control; n = 3; blue bars) and TBS (TBS276∗ and TBS332; pooled; n = 5; orange bars) are shown.
(E) Graphical representation of the fold change in luciferase activation when Shh-LIGHT2 control cells (n = 8; blue bars) or cells expressing the mutated form GFS-SALL1c.826C>T (SALL1276; n = 8; orange bars) were either treated (+) or not (−) with purmorphamine.
All graphs represent the mean and SEM. p values were calculated with a two-tailed unpaired Student’s t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.