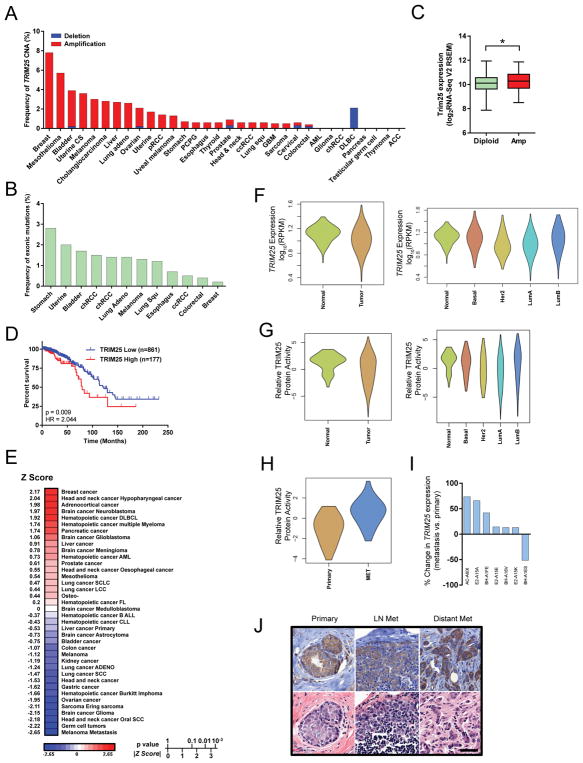
Figure 7.
TRIM25 is frequently amplified in breast cancer and is associated with poor overall survival. (A) Frequency of TRIM25 copy number alterations (CNA) in 32 cancer types from the TCGA. CS, cell carcinoma; pRCC, papillary renal cell carcinoma; PCPG, pheochromocytoma and paraganglioma; ccRCC, clear cell renal cell carcinoma; GBM, glioblastoma multiforme; AML, acute myleiod leukemia; chRCC, chromophobe renal cell carcinoma; DLBC, diffuse large B-cell lymphoma; ACC, adenoid cystic carcinoma. (B) Mutation frequency of TRIM25 in 32 cancer types from the TCGA. Cancers not represented on graph have no known mutations. (C) TRIM25 expression in TRIM25 diploid vs.TRIM25 amplified tumors (TCGA). (D) Kaplan-Meier survival analysis based on TRIM25 expression (RNA-seq v2 RSEM) from 1,038 breast cancer patients from TCGA. TRIM25 ‘High’ is defined by a z-score>1, TRIM25 ‘Low’ is all other patients. HR, hazard ratio. (E) PREdiction of Clinical Outcomes from Genomic profiles (PRECOG) analysis of TRIM25. (F) TRIM25 expression in breast tumors vs normal breast tissue. (G) Predictive TRIM25 relative activity in breast tumors vs normal breast tissue. (H) Predictive TRIM25 relative activity in primary breast cancer tumors vs. metastases. (I) TRIM25 expression in primary breast cancer tumors vs. matched metastases (TCGA IDs indicated). (J) Immunohistochemistry of TRIM25 in primary breast tumor, lymph-node and distant (bone) metastases. Scale bar represents 50μm.