FIGURE 4.
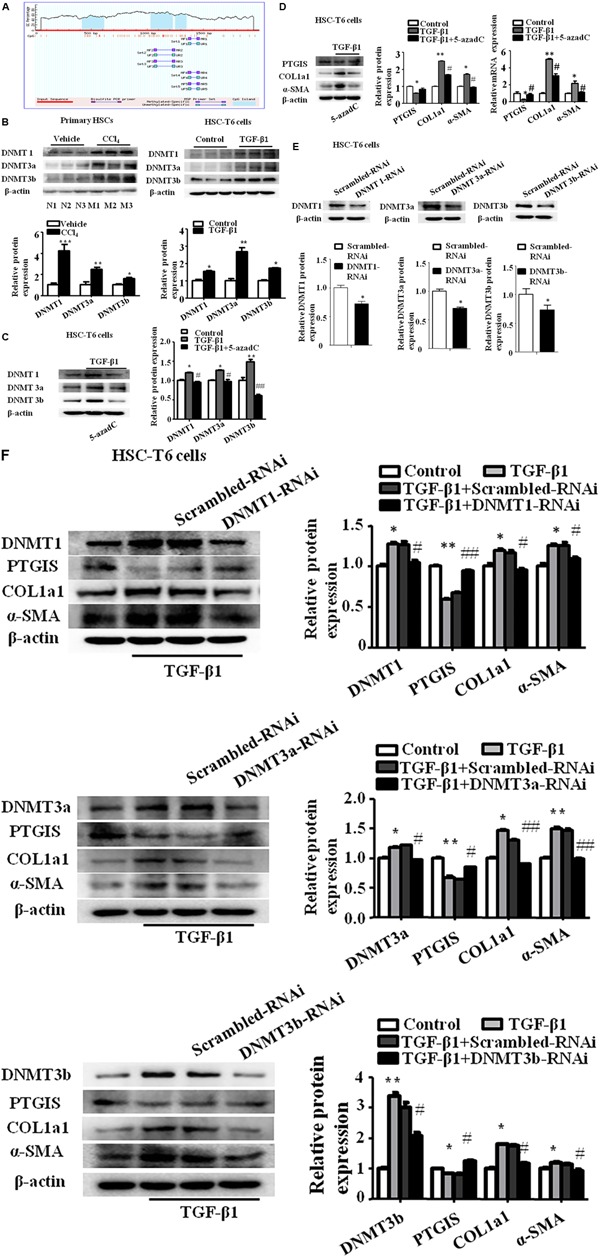
The expression of PTGIS was associated with DNA hypermethylation in activated HSC-T6 cells. (A) The CpG island in PTGIS gene were predicted. (B) The protein levels of DNMT1, DNMT3a, and DNMT3b in primary HSCs isolated from fibrotic livers and HSC-T6 cells were detected by western blot analysis, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. vehicle group or control group. (C) The protein levels of DNMT1, DNMT3a and DNMT3b in control, model and TGF-β1 + 5-azadC groups, HSC-T6 cells were treated with 5-azadC (2 μM) 6 h before added TGF-β1(10 ng/mL) for 24 h, ∗p < 0.05, ∗∗p < 0.01 vs. control group, #p < 0.05, ##p < 0.01 vs. TGF-β1-treated group. (D) The protein and mRNA levels of PTGIS, COL1a1, and α-SMA were measured by western blot and RT-qPCR, ∗P < 0.05, ∗∗P < 0.01 vs. control group, #p < 0.05 vs. TGF-β1 group. (E) The efficiency of DNMT1, DNMT3a and DNMT3b lose-expression in HSC-T6 cells were detected by western blot, ∗p < 0.05 vs. Scrambled-RNAi group. (F) The protein levels of DNMT1, DNMT3a, DNMT3b, PTGIS, COL1a1 and α-SMA were detected via western blot analysis, ∗P < 0.1, ∗∗P < 0.05 vs. control group, #P < 0.1, ##P < 0.05 vs. TGF-β1 + Scrambled-RNAi group.
