Figure 1. Predicting coevolved residues in the group II chaperonins.
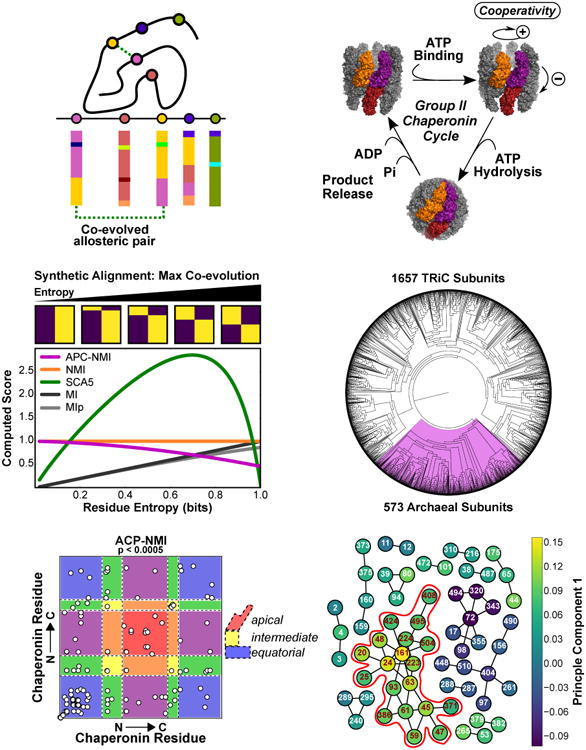
(A) Coevolution prediction theory. An important allosteric interaction exists between two residues. A sequence alignment for this protein family reveals a pattern of coevolution between these positions.
(B) Group II chaperonins undergo a nucleotide dependent conformational cycle. Three subunits are highlighted to show intra- and inter-ring pairs. For simplicity, proposed cooperative transitions are schematically indicated in the symmetric complex, however asymmetric intermediates have been proposed [47,62].
(C) The entropy dependence of various measures for two residue alignments with two residue types in the case of maximal covariation. Sample alignments are shown above the plot and move from nearly perfect conservation (left) to 1.0 bit of entropy (right). The pairwise score is computed for the indicated measure as a function of shannon entropy. We do not evaluate APC-NMI at zero entropy as it is undefined there.
(D) A phylogenetic tree of the group II chaperonin sequences found in the CpnDB. Residues with very low entropies are pruned from our alignments before analysis.
(E) Predicted covarying positions identified via empirical thresholding of the APC-NMI scores, p values were generated by bootstrapping the distribution of pairwise APC-NMI scores. Background colored according to the domain architecture of the chaperonin subunit. Blue: equatorial; Yellow, intermediate; Red apical domains. Colors blended accordingly for couples between domains.
(F) Interconnected networks formed from coupled residue pairs in F; colored according to the first component of the APC-NMI matrix decomposition. (Red outline) The largest interconnected network can also be seen to strongly correlate with the first component.
