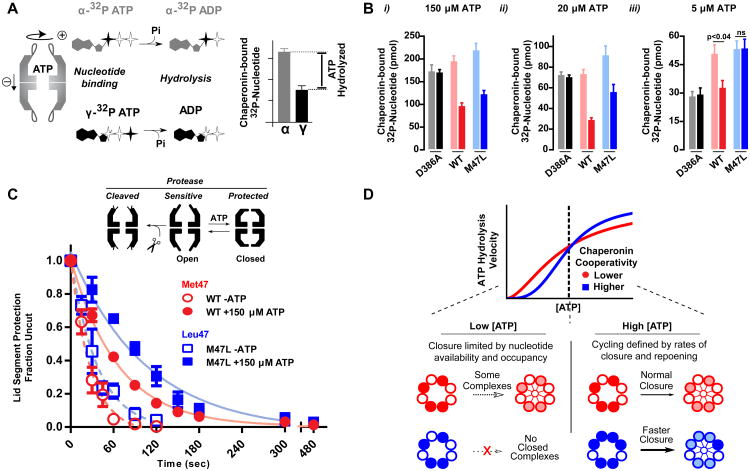
Figure 4. Chaperonin cycling is defined by nucleotide occupancy.
(A) A nucleotide recovery assay utilizing both α- and γ- labeled 32P-ATP allows for differentiating between the phosphorylation state of bound nucleotide and calculating the amount of hydrolyzed nucleotide.
(B) Nucleotide bound to the chaperonin at the indicated concentrations was separated utilizing HA-nitrocellulose filter membranes prior to liquid scintillation. The hydrolysis deficient mutant, D386A, is shown as a control. Data shown as the mean ± s.e.m. of three independent samples. A t-test was performed
(C) State of the chaperonin monitored by protease sensitivity. Complexes were incubated with 40 ng/uL Proteinase K at indicated nucleotide conditions. Fraction uncut determined by SYPRO Ruby fluorescence after separation by SDS-PAGE. Data is the mean ± s.e.m of three independent replicates.
(D) Allosteric differences between WT and M47L reveal how the folding cycle of the group II chaperonins is regulated under different nucleotide concentration regimes.