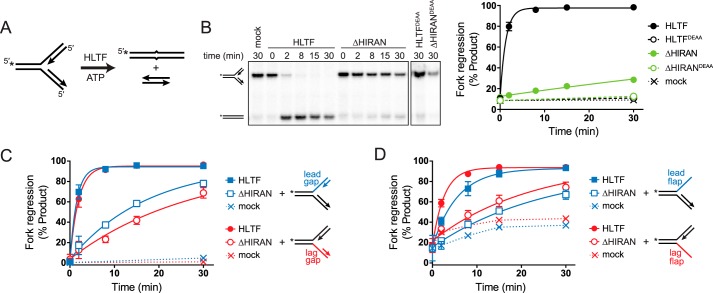
Figure 5.
HIRAN is not required for regression of forks containing ssDNA at the junction. A, schematic of fork regression assay. A model fork substrate containing a two-nucleotide mismatch at the junction to prevent spontaneous branch migration is incubated with HLTF and ATP to yield annealed template and nascent duplexes. 32P labels are indicated by asterisks. B, representative native gel of time-dependent fork regression by HLTF and ΔHIRAN against a fork substrate with fully paired nascent arms. Endpoints of reactions containing ATPase-dead HLTFDEAA or ΔHIRANDEAA proteins, or of a mock reaction with no protein are shown as negative controls. The plot shows quantitation of three experiments (mean ± S.D.). Data were fit by a single exponential function. C and D, quantitation of three time-dependent fork regression reactions (mean ± S.D.) with HLTF and ΔHIRAN using forks in which the leading or lagging nascent strands have been shortened (C) or removed (D). Mock reactions contain no enzyme. Representative gels for panels C and D are shown in Fig. S3.