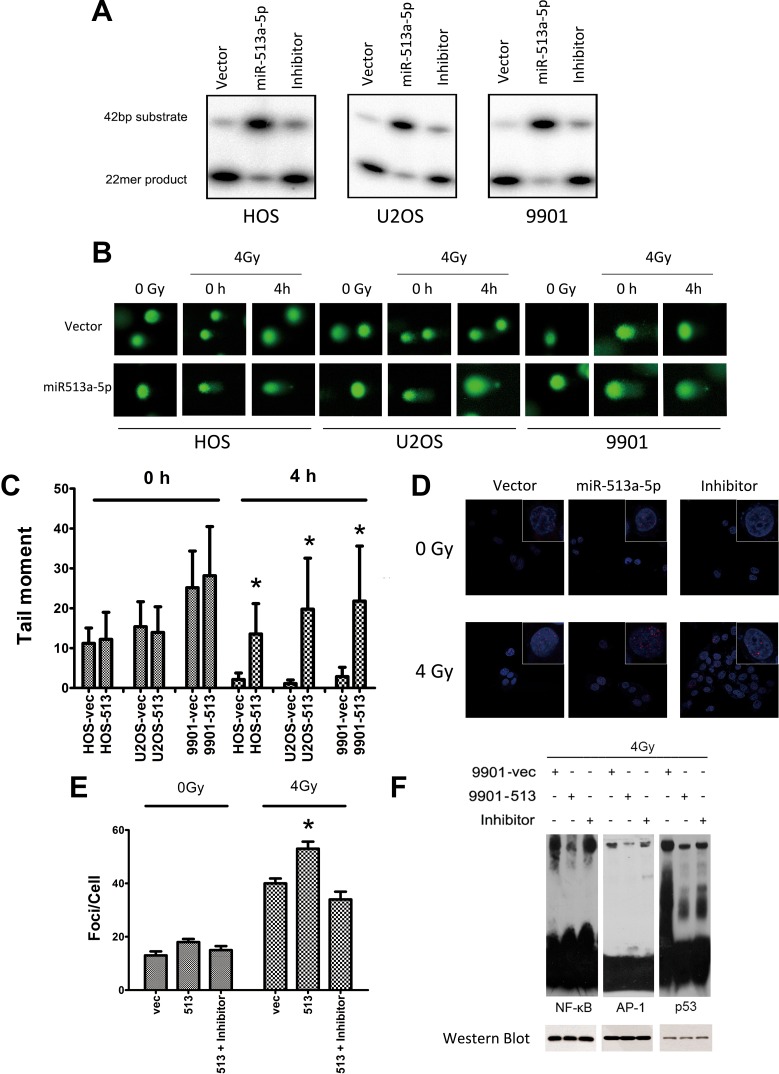
Figure 4. miR-513a-5p decreases DNA repair and redox activity of APE1 in osteosarcoma cells.
(A) AP endonuclease activities of APE1 protein in the control cell line, stably-expressing miR-513a-5p cells, and inhibitor treated HOS, U2OS and 9901 cells were detected by abasic site incision assay. (B) Osteosarcoma cells with stable miR-513a-5p expression and control cells received 4 Gy X-ray or sham (0Gy) irradiation, and were detected by alkaline comet assay immediately (0 h) or after recovery for 4 hours (4 h) to evaluate the overall DNA repair activity of single strand breaks. Representative images of the comet assays are shown. (C) Statistical analysis of comet assay and asterisk (*) indicates that the difference between the marked group and the vector-transfected group is statistically significant (p < 0.01). (D) γ-H2AX foci were visualized (red) at 4 hours post irradiation or sham by immunofluorescent assay. Nuclei were stained by DAPI as shown in blue. Only the representative images from each group are shown in the merged pattern. (E) Statistical analysis of γ-H2AX foci staining counts and asterisk (*) indicates that the difference between the marked group and the vector-transfected group is statistically significant (p < 0.01). The impact of miR-513a-5p level on the redox activity of APE1 was analyzed by EMSA using the probes containing the binding sequences of NF-κB, p53 and AP-1, which are transcription factors regulated by APE1 redox activity (F). The cells were harvested at 24 hour after 4 Gy X-ray irradiation. To verify the loading of nuclear extracts in each reaction was equal, Western blot against NF-κB (p50), p53 and AP-1 were shown in the bottom panel.