Figure 2.
Integration of Deep Transcriptomic and Proteomics Data
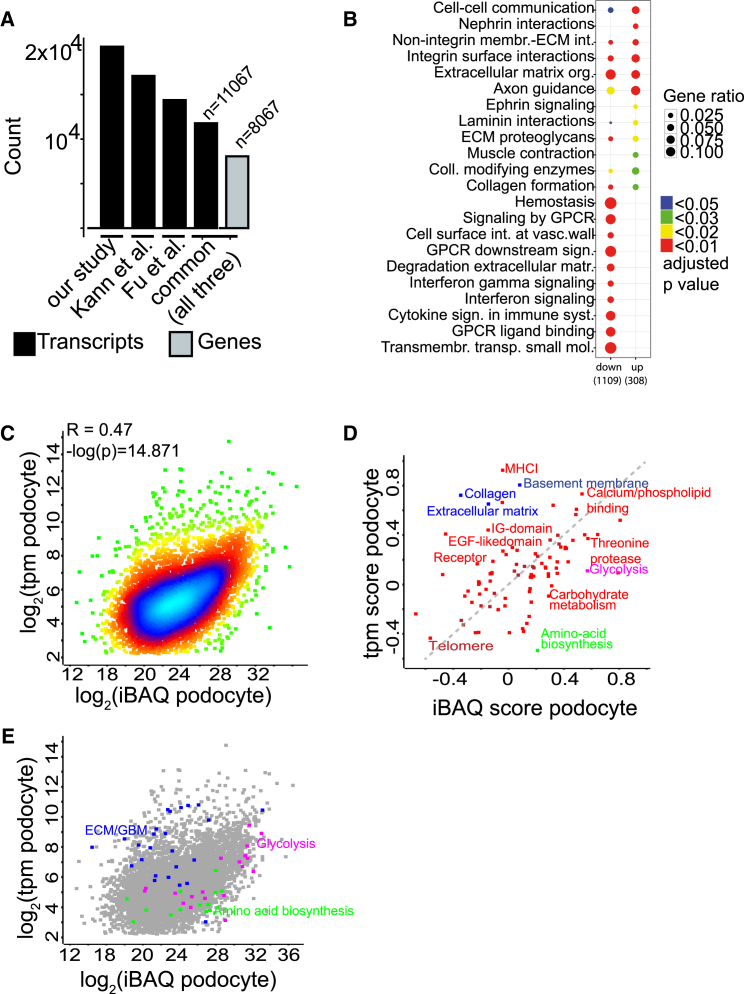
(A) Comparison of this and other recent transcriptomic (tpm) studies using mRNA-seq. All datasets were processed with the same bioinformatics pipeline as indicated in the Experimental Procedures.
(B) Reactome analysis of transcripts enriched in podocytes (up) and de-enriched in podocytes (down).
(C) Scatterplot demonstrating correlation between proteomic (iBAQ) and (tpm) copy numbers in podocytes (Pearson’s R = 0.47, log(p) = 14.8). The density of individual proteins is color-coded (blue, high density; green, low density).
(D) 2D Uniprot keyword analysis of podocyte transcript and protein copy numbers identifies significantly changed Uniprot keywords with an especially high increase in either omic dataset (permutation-based FDR < 0.05). The dashed gray line (y = x) indicates annotations equally represented in both omic datasets.
(E) Distribution of individual Uniprot keyword members in the original scatterplot (analog to C).