Figure 3.
Determination of Podocyte-Enriched Proteins Uncovers Druggable Targets and Candidate Genes for Podocyte Function
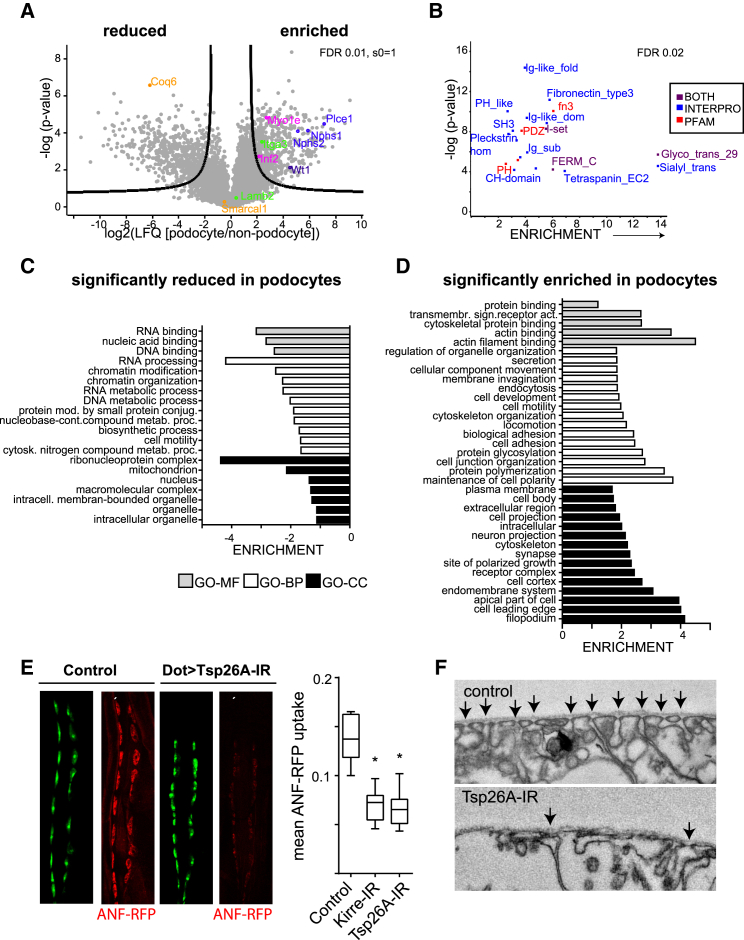
(A) Volcano plot of protein quantification of podocytes over non-podocytes. The logarithmic fold change of label-free quantification intensities (LFQs) is plotted versus the negative decadic logarithm of the p value. 551 proteins pass the criteria for significant podocyte enrichment (significance analysis of microarrays [SAM] analysis, s0 = 1, FDR = 0.01).
(B) Enrichment analysis of protein domains in podocytes. Protein domains (PFAM and INTERPO annotation) were tested for enrichment within the podocyte-enriched proteins over the proteins not significantly enriched. –log(p value) of every significantly enriched protein domain is plotted against fold enrichment. Fisher’s exact test, FDR < 0.02.
(C) GO terms overrepresented in the 551 podocyte-enriched proteins compared with non-podocyte specific proteins. Fold enrichment of GO terms representing molecular function (GOMF, gray bars), cellular component (GOCC, black bars), and biological process (GOBP, white bars). All GO terms are statistically enriched in the dataset compared with non-podocyte proteins. Fishers exact test, FDR < 0.05.
(D) Significantly de-enriched GO terms in the 551 podocyte-specific proteins (the same statistical criteria and color code as in C).
(E) CLSM micrographs of pericardial nephrocytes at second larval stage. RNAi-mediated knockdown of Tsp26A significantly reduced ANF-RFP (red) uptake, suggesting that Tsp26A is required for pericardial nephrocyte (green) function. The ANF-RFP uptake is significantly reduced by the knockdown of Tsp26A (p value < 0.05)
(F) Ultrastructural alteration of Tsp26A knockdown nephrocytes. Arrows indicate nephrocyte cell contacts.