Figure 4.
Comparison of the Deep Native (Isolated from Living Mice) and Cultured Podocyte Proteome Enables Functional Analysis of Cell Culture-Induced Proteome Artefacts
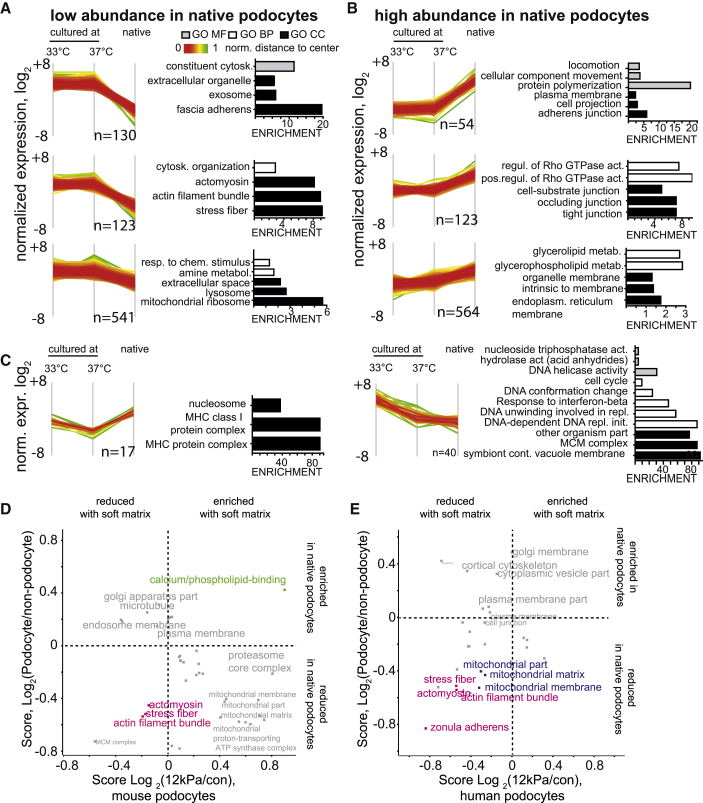
(A) Protein clusters were defined based on normalized intensities as depicted in Figure S4C–D. Shown are clusters that have a higher intensity in cultured podocytes compared with the native podocyte proteome. Distance to the mean is color-coded in each cluster. GO terms significantly overrepresented in each cluster are depicted (Fishers exact test, p values corrected with FDR < 0.05).
(B) Three clusters have a lower intensity (and abundance) in the podocyte proteome compared with the cultured podocyte proteome. GO terms significantly overrepresented in each cluster are depicted.
(C) Two clusters have different intensities in undifferentiated and differentiated podocytes.
(D) Mouse podocytes were seeded on matrices with 12 kPa, and the proteome was compared with control cell culture dishes. See Figure S5 for details regarding the dataset. The 2D GO enrichment score of fold change on soft matrix (log2 12 kPa/control) cell culture is plotted against the score for podocyte-enrichment (log2(podocyte/non-podocyte)) (Figure 3). The data demonstrate a significant decrease of stress fiber proteins under both conditions (soft versus stiff matrix and native podocyte versus non-podocyte). Significantly changed terms are plotted (FDR < 0.05)
(E) Human podocytes were seeded on matrices with 12 kPa, and the proteome was compared with control cell culture dishes. See Figure S5 for details regarding the dataset.