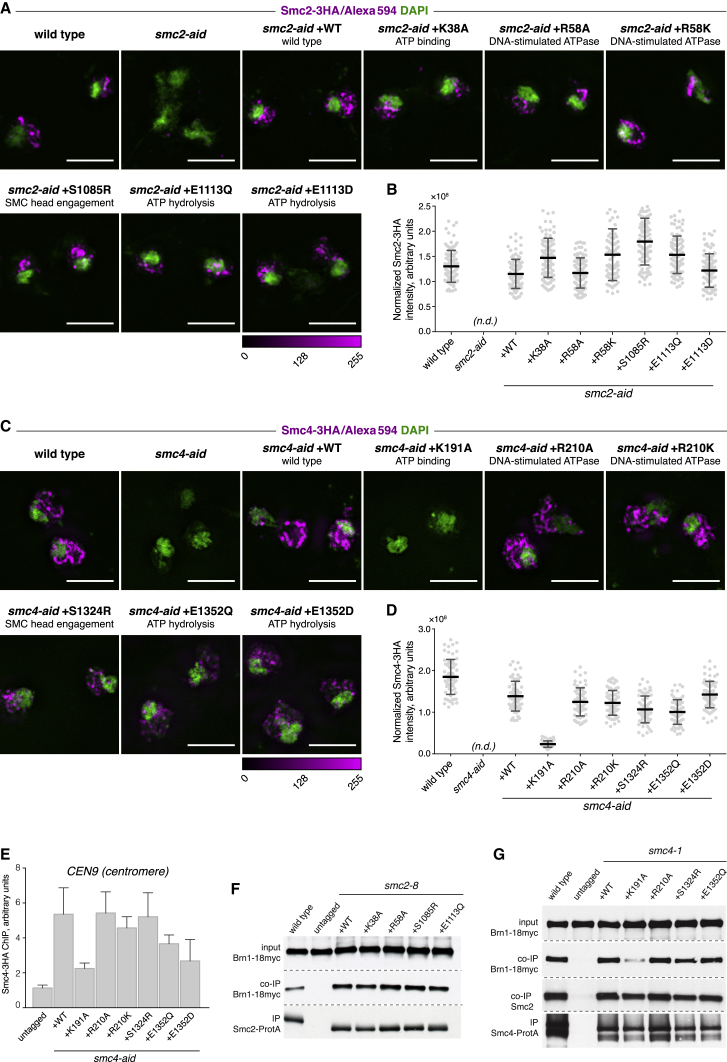
Figure 1.
ATPase-Independent Chromosome Binding of Condensin
(A) Chromosome spreads of wild-type or smc2-aid cells in metaphase expressing ectopic 3HA-tagged wild-type or ATPase mutant Smc2. Cells were synchronized in G1 with α factor and released into a nocodazole-induced metaphase arrest. Chromosome spreads were stained with DAPI and anti-HA/Alexa Fluor 594 antibodies. Scale bars represent 4 μm.
(B) Quantification of the Smc2-3HA staining intensities in (A), normalized by cellular expression levels assessed by immunoblotting. Error bars represent mean ± SD (n ≥ 92).
(C) Chromosome spreads of wild-type or smc4-aid cells in metaphase expressing ectopic 3HA-tagged wild-type or ATPase mutants of Smc4, as in (A).
(D) Quantification of the Smc4-3HA staining intensities in (C), normalized by cellular expression levels assessed by immunoblotting. Error bars represent mean ± SD (n ≥ 63).
(E) Chromatin immunoprecipitation (ChIP)-qPCR signal of Smc4-3HA at CEN9, normalized to a negative binding site, in smc4-aid cells in metaphase expressing ectopic wild-type or ATPase mutants of Smc4. Error bars represent mean ± SEM (n = 3).
(F) Coimmunoprecipitation of Brn1-18myc with ATPase mutants of Smc2, as assessed following their immunoprecipitation by means of a Protein A tag in a temperature-sensitive smc2-8 background.
(G) Coimmunoprecipitation of Brn1-18myc with ATPase mutants of Smc4, as assessed following their immunoprecipitation by means of a Protein A tag in a temperature-sensitive smc4-1 background.
See also Figure S1 for a viability assay of cells harboring condensin SMC ATPase mutations; Figure S2 for confirmation of cell-cycle synchrony, raw staining intensities, and protein expression levels used for normalization; and Figure S3 for ChIP-qPCR analyses at additional loci.