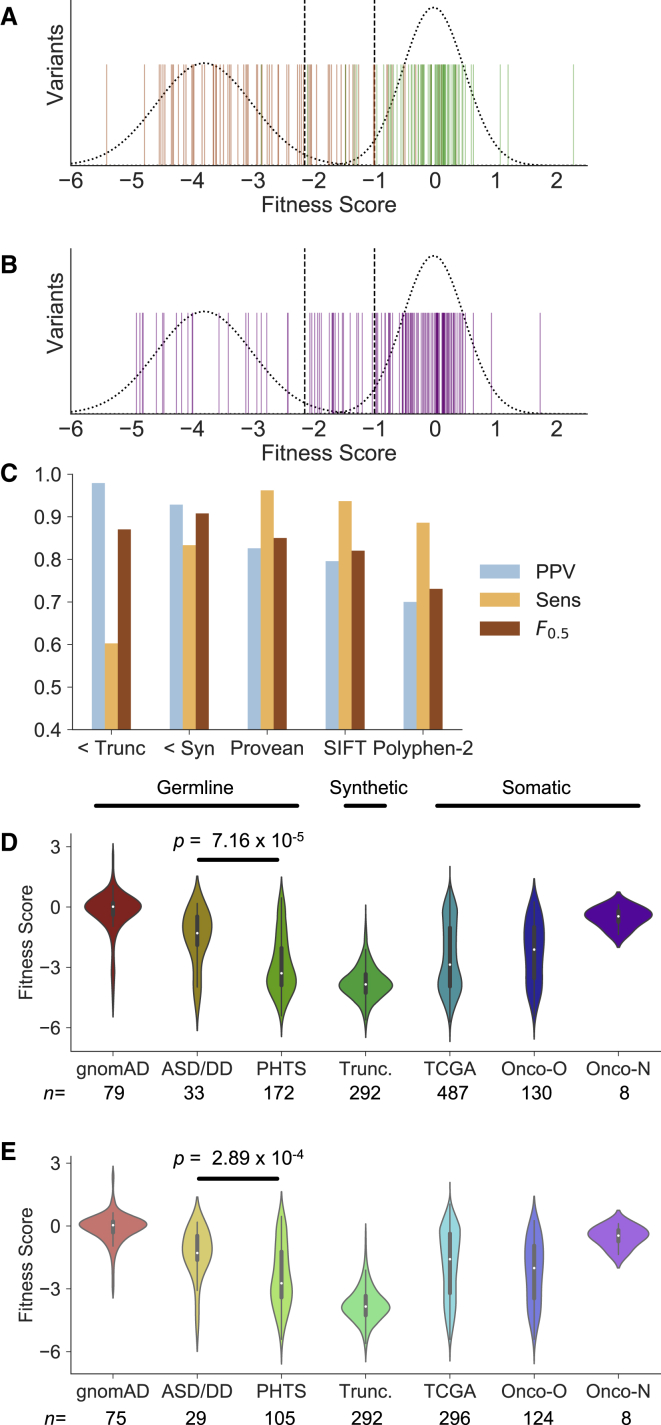
Figure 4.
Fitness Scores Discriminate between Likely Pathogenic and Benign Variants and Support Genotype-Phenotype Relationships
(A) Fitness scores for missense variants considered pathogenic or likely pathogenic in ClinVar (orange) and putatively benign variants from gnomAD (green). Dashed curves represent truncation (left) and synonymous (right) distributions. Dashed lines at −2.15 and −1 represent the approximate 95% two-tailed distribution of truncations (before regulatory tail) and synonymous mutations, respectively.
(B) Fitness scores of ClinVar VUS (purple), with truncation and synonymous distributions and 95% limits.
(C) To test the ability of fitness scores to discriminate likely pathogenic from likely benign missense mutations, we calculated positive predictive value (PPV), sensitivity, and F0.5 scores for our fitness scores (“<Trunc” represents the threshold at −2.15, “<Syn” represents the threshold at −1). In these tests, a true positive represented a ClinVar pathogenic allele having a fitness score less than or equal to the threshold. We compared the performance of the fitness scores at these two thresholds with in silico pathogenicity predictors for missense mutations using their default thresholds (Material and Methods).
(D) Fitness scores of all curated mutations associated with the indicated phenotype (Material and Methods).
(E) Fitness scores of only curated missense mutations associated with the indicated phenotype (programmed truncations also shown for clarity). Abbreviations: ASD/DD, autism spectrum disorder/developmental delay; PHTS, PTEN hamartoma tumor syndrome; TCGA, The Cancer Genome Atlas; Onco-O, OncoKB mutations considered “oncogenic” or “likely oncogenic;” Onco-N, OncoKB mutations considered “likely neutral.”