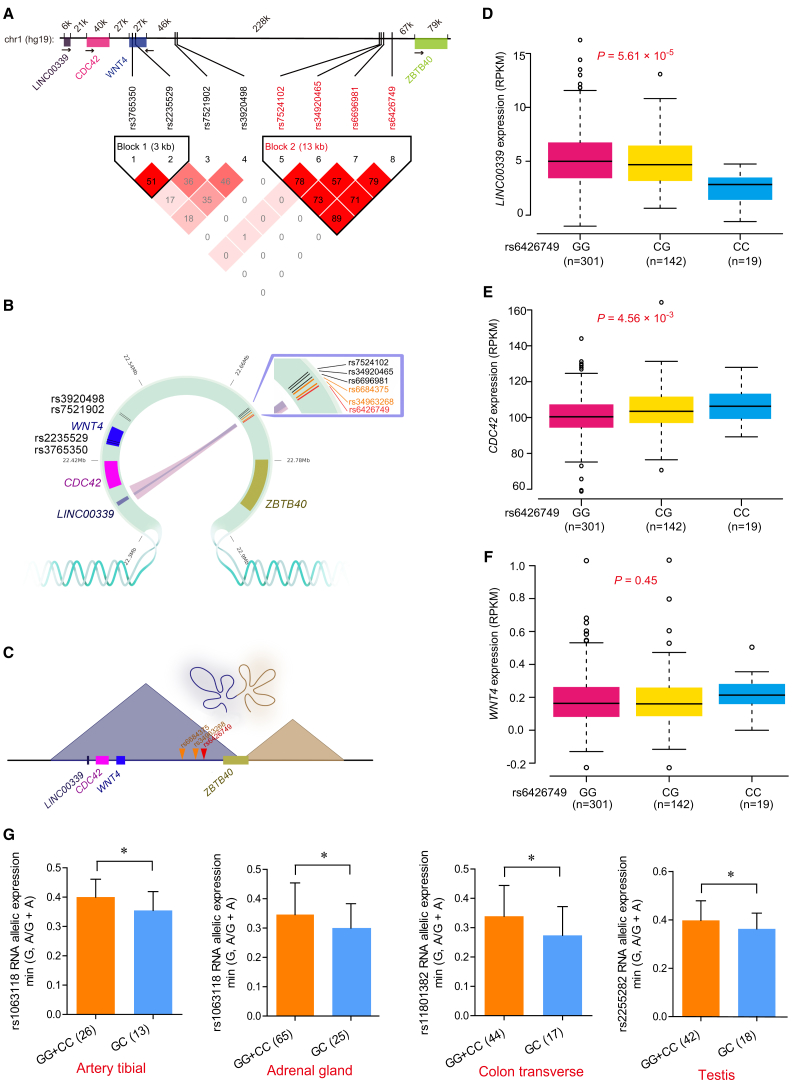
Figure 2.
Integrating Analyses Indicate the Long-Range Interaction between rs6426749 and LINC00339
(A) LD blocks for eight BMD SNPs. The upper bar shows genomic positions for eight BMD SNPs in 1p36.12 and nearby genes, with distance between genes and (or) SNPs displayed above (kb). The bottom inverted triangle shows the LD blocks for eight BMD SNPs at 1p36.12, with each diamond representing the r2 measure of LD using standard color scheme, where the darker shades of red represent greater values.
(B) Hi-C interactions between eQTLs and promoters of target genes, and different color of lines indicated different Hi-C dataset (pink: Hi-C data on IMR90 cells from 4DGenome;25 blue: DNase Hi-C data on H1-hESC cells30). SNP rs6426749 overlapped with Hi-C regions is labeled in red. Another two SNPs in strong LD with rs6426749 within the same Hi-C interaction regions are labeled in orange.
(C) The loop between rs6426749 and LINC00339 is located within a 600 kb topologically associated domain (TAD) in IMR90 cells.
(D–F) Boxplot for LINC00339 (D) or CDC42 (E) or WNT4 (F) expression in samples with different genotypes of rs6426749 (GG, CG, CC) taken from 462 LCLs samples.54 Sample counts are shown.
(G) Allele-specific expression (ASE) analysis between rs6426749 and LINC00339, using monoallelic gene expression data from GTEx.40 Four significant tissues (p < 0.05) are shown. The horizontal axis represents individuals with homozygous or homozygous genotypes for rs6426749. The vertical axis represents the exonic SNP chosen as a measurement of allelic expression of LINC00339. Error bars, SD; ∗p < 0.05 as determined by Wilcoxon rank sum test.