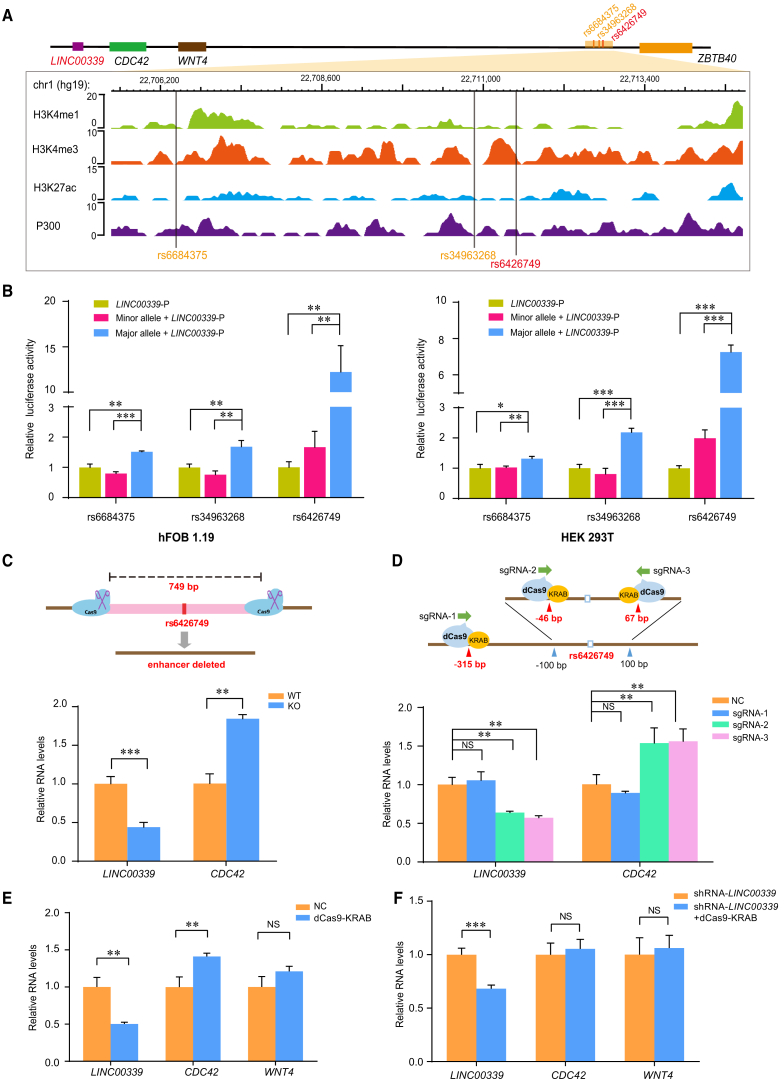
Figure 3.
The Region Containing rs6426749 Acts as Strong Allele-Specific Enhancer on the LINC00339 Promoter
(A) Epigenetic annotation for region surrounding rs6684375, rs34963268, and rs6426749 in osteoblast cells. The data are obtained from ENCODE Project taken from WashU EpiGenome Browser, including active histone modification (H3k4me1, H3K4me3, H3k27ac) as well as acetyltransferase (P300).
(B) The dual-luciferase assay for LINC00339 promoter (LINC00339-P) containing rs6684375, rs34963268, or rs6426749 locus with either the major or minor allele, or individual LINC00339-P was measured in hFOB 1.19 cells or HEK293T cells. The individual LINC00339-P was used as baseline control. Luciferase signal was normalized to Renilla signal. Error bars, SD. n ≥ 3. ∗∗p < 0.01, ∗∗∗p < 0.001 as determined by an unpaired, two-tailed Student’s t test.
(C) Comparison of LINC00339 and CDC42 expression between rs6426749 region deleted hFOB 1.19 cells (KO) mediated by CRISPR/Cas9 and normal cells (WT).
(D) Comparison of LINC00339 and CDC42 expression between rs6426749-locus repressed hFOB 1.19 cells using dCas9-KRAB and normal cells (NC, negative control). One distal sgRNA (sgRNA-1) and two proximal sgRNAs (sgRNA-2, sgRNA-3) were designed.
(E) Effect of rs6426749-locus repression in hFOB 1.19 cells using dCas9-KRAB (sgRNA-3) on LINC00339, CDC42, and WNT4 expression.
(F) RT-qPCR for LINC00339, CDC42, and WNT4 expressions in hFOB 1.19 cells after silencing of both LINC00339 using shRNA and rs6426749-locus using dCas9-KRAB (blue) as compared with LINC00339 silenced cells (orange), respectively. Error bars, SD. n ≥ 3. NS: not significant, ∗∗p < 0.01, ∗∗∗p < 0.001 as determined by an unpaired, two-tailed Student’s t test.