Abstract
Loss of expression of ACTN3, due to homozygosity of the common null polymorphism (p.Arg577X), is underrepresented in elite sprint/power athletes and has been associated with reduced muscle mass and strength in humans and mice. To investigate ACTN3 gene dosage in performance and whether expression could enhance muscle force, we performed meta-analysis and expression studies. Our general meta-analysis using a Bayesian random effects model in elite sprint/power athlete cohorts demonstrated a consistent homozygous-group effect across studies (per allele OR = 1.4, 95% CI 1.3–1.6) but substantial heterogeneity in heterozygotes. In mouse muscle, rAAV-mediated gene transfer overexpressed and rescued α-actinin-3 expression. Contrary to expectation, in vivo “doping” of ACTN3 at low to moderate doses demonstrated an absence of any change in function. At high doses, ACTN3 is toxic and detrimental to force generation, to demonstrate gene doping with supposedly performance-enhancing isoforms of sarcomeric proteins can be detrimental for muscle function. Restoration of α-actinin-3 did not enhance muscle mass but highlighted the primary role of α-actinin-3 in modulating muscle metabolism with altered fatiguability. This is the first study to express a Z-disk protein in healthy skeletal muscle and measure the in vivo effect. The sensitive balance of the sarcomeric proteins and muscle function has relevant implications in areas of gene doping in performance and therapy for neuromuscular disease.
Keywords: ACTN3, actinin-3, alpha actinin 3, gene doping, muscle, rAAV, Z-line, Z-disk, skeletal muscle, fast fibers
Introduction
The ACTN3 gene encodes for α-actinin-3, an actin-binding protein that is specifically expressed in fast skeletal muscle fibers (all fast glycolytic type 2X fibers and 50% of fast oxidative type 2A fibers).1 The common ACTN3 polymorphism (Arg577X) (rs1815739) is one of the most highly replicated genetic associations in human muscle performance. Homozygosity of the 577X allele (577XX) occurs in 20% of the global population2 and results in complete loss of function (absence) of α-actinin-3. Association studies in sprint/power performance have consistently demonstrated reduced 577X allele frequency in elite athletes, suggesting that the expression of α-actinin-3 (or, presence of 577R allele) plays an integral role for optimal generation of explosive muscle power. No other common variant or study has been reproducibly associated with elite sprint/power performance.3
The effect of ACTN3 Arg577X on function has also been examined in various non-athlete populations. Absence of α-actinin-3 has been associated with loss of muscle power4 and strength5 reduction of bone and muscle mass6, 7 as well as susceptibility to injury.8 Other studies have also shown that the absence of α-actinin-3 is linked to increased risk of falls in the elderly,9 earlier onset and poorer prognosis among individuals with Pompe disease (MIM: 232300),10 and reduced muscle strength in boys with Duchenne muscular dystrophy (MIM: 310200).11 Mechanistic studies on healthy human muscle reported a slower, more oxidative muscle profile in people carrying two copies of the null 577X allele, with some evidence for shifts in fiber-type proportions,12, 13 increased muscle glycogen,14, 15 calcineurin signaling,16 and expression of structural and oxidative signaling proteins.17 Collectively, these studies identify ACTN3 as an important influence upon muscle attributes in the context of elite performance, healthy individuals, and among those contending with heritable and acquired conditions of compromised muscle function.
Phenotypic analyses of the Actn3 knockout (KO) mouse model recapitulated the human 577XX phenotypes, with Actn3 KO mice showing reductions in muscle strength, increased endurance capacity,18 reduced fast fiber size, shifts in fast fiber metabolism toward oxidative metabolism,14, 18 enhanced calcineurin signaling, and increased response to endurance exercise training compared to wild-type (WT) mice.16 Similar to 577XX human findings, impacts on muscle function are life-long, with aged Actn3 KO mice showed reduced muscle strength, muscle mass, and fast fiber size compared to WT mice.19 The expression pattern of Actn3 in the mouse muscle remains similar to the human (Actn3 is expressed in fast 2B and 2X fibers but not type 2A or I), but the impact of Actn3 deficiency is likely accentuated due to the higher proportion of fast fibers in a mouse compared to a human muscle.1, 20 For this reason, human association studies remain critical to ascertain the effect of ACTN3 deficiency.
One of the most studied and replicated human association study results is the detrimental effect of α-actinin-3 deficiency (577XX) on elite sprint/power muscle performance. However, the nature of the genetic model that best explains the association—for example, an α-actinin-3 gene dosage effect (RR > RX > XX), a recessive effect (XX versus RR/RX), or a dominant effect (RR versus RX/XX)—has not been properly explored.4 Physiological investigations (phenotypes and expression data) in the Actn3+/− heterozygous (HET) and homozygous null (KO) mice supported the use of an additive model when testing ACTN3 genotype associations.21 The additive model (KO > HET > WT) best explained relative muscle force (percentage of maximum) in response to fatigue, the reduction in 2B fiber size, and increases in calcineurin signaling and oxidative metabolism.21 Expression of Actn3 in HET mouse muscles was lower relative to WT and were compensated by upregulation of α-actinin-2 in a dose-dependent, genotype manner. Similarly, Z-disk proteins such as ZASP, mitochondrial proteins (COX IV, porin), and calcineurin signaling (as shown by downstream RCAN1-4 expression) were at intermediate levels in HET mice relative to WT and KO.21
Collectively, the results reported to date suggest that restoring α-actinin-3 expression may increase muscle mass and force-producing capacity in α-actinin-3-deficient skeletal muscle. We and others have previously shown that recombinant adeno-associated viral vectors (rAAV) can efficiently transfer genes to mature mouse muscles, resulting in long-term gene expression.16, 22 Given the ACTN3 Arg577X association with human muscle performance and muscle traits throughout life, augmenting expression of α-actinin-3 in skeletal muscle could further enhance muscle power and sprint performance in individuals already expressing α-actinin-3 (RR, RX). Additionally, replacement of α-actinin-3 in muscles of ACTN3 577XX individuals could improve muscle power performance with beneficial consequences for prognosis in regards to muscle-wasting disorders and mitigating the risks of falling in the elderly.23
In this study, we examined the potential of ACTN3 overexpression in mature skeletal muscle to enhance muscle strength, mass, and the susceptibility/resistance to fatigue in the course of repeated stimulation. A general meta-analysis was first performed using data from published, independent studies of ACTN3 Arg577X to determine what type of genetic model best explains the association with elite sprint/power performance. Using the Actn3 WT and KO mouse models, we further investigated the functional and morphological effects of post-natal manipulation of α-actinin-3 dosage by overexpressing sarcomeric α-actinin-3 in mature skeletal muscle via rAAV-mediated gene transfer. We hypothesized that altering skeletal muscle α-actinin-3 expression would modify muscle function in a dose-dependent fashion.
Material and Methods
Study Design
Regarding systematic meta-analysis, refer to Supplemental Material and Methods.
For mouse studies, sample sizes (n > 5) were selected based on previous Actn3 KO and WT studies.11, 21, 24 All experiments were replicated in independent groups of mice or using different muscles (TA and EDL). Functional testing was performed by investigators that were blinded to genotype and treatment. All data were collected from male mice and age-matched littermates.
Mice Housing Conditions
Mice were from C57BL/6 (N12-17) genetic backgrounds aged between 6 and 24 weeks. One group of WT mice were from a house colony of C57BL/6 (N70). Mice were housed under the same conditions and were fed meat-free mouse pellets and water ad libitum and were maintained in a 12:12 hr cycle of light and dark at ambient room temperature (∼22°C). All experiments were approved by the Animal Care and Ethics Committee of the Children’s Medical Research Institute (CMRI approval ID: K190) and the Murdoch Children’s Research Institute (MCRI approval ID: A761).
rAAV Vector and Delivery
The generation of recombinant adeno-associated viral (rAAV6) vectors was previously described.16 For intramuscular injections, mice were anesthetized (3.5% isoflurane in oxygen from a precision vaporizer) and given temgesic (0.05 mg/kg) for pain management. Hamilton syringes (32 gauge) (Hamilton) were used to deliver 30 μL of rAAV-ACTN3 or rAAV-MCS (control empty vector) diluted with of HBSS to required dose (5E8-1E11 vector genomes). Vectors were injected into the anterior compartment of the lower limb, targeting the TA and EDL.
Muscle Physiology
Ex vivo EDL: Mice were sacrificed by cervical dislocation and the EDL was isolated to determine muscle force and recovery from fatigue. All experiments were carried out at room temperature (∼22°C–24°C) using techniques previously described.18, 24 In situ TA: Force and fatigue was performed using the Aurora Scientific Dual Mode Lever System with supplied software (DMC5 4.5, DMA) and carried out at 37°C as previously described.11 Notes on both protocols are supplied in the Supplemental Material and Methods.
Immunoblotting
Immunoblotting was performed as previously described.11 Briefly, 10-μm frozen muscle sections were homogenized in 4% SDS lysis buffer and assessed for total protein concentration using the Pierce BCA protein assay kit (ThermoFisher Scientific). Proteins were separated by SDS-PAGE using precast minigels (Invitrogen) or criterion gels (Biorad), then transferred to polyvinylidene fluoride membranes (PVDF, Millipore). These were blocked with 5% skim milk in 1× PBST, probed with probed antibodies (Supplemental Material and Methods) overnight and secondary antibodies at room temperature for 2 hr, then developed with ECL reagents (Amersham Biosciences). Membranes were washed, developed, and imaged by Image Quant (GE Healthcare). Densitometry was performed using ImageJ image processing software (NIH) and quantified using the area under the curve. This was normalized to myosin and actin expression, obtained from PVDF membranes stained with Coomassie brilliant blue (1610436, Sigma-Aldrich).
Histology and Immunofluorescence
Haematoxylin and eosin and SDH histological stains were performed on 8-μm muscle sections as previously described.18 For immunofluorescence, 8-μm sections were blocked with 2% BSA or affinipure goat α-mouse Fab fragment (Jackson ImmunoResearch) before overnight primary incubation (antibodies are in Supplemental Material and Methods) at 4°C. Sections were washed with 1× PBS. Secondary incubation was performed in combination with Hoechst staining at room temperature for 1 hr. Fiber-typing was performed as previously described.25 Sections were imaged on the Mirax slide scanner (ZEISS), V-Slide Scanner (MetaSystems), or the confocal microscope (Zeiss LSM 780).
Statistics for Experimental Studies
Data results (sample size) are presented as individual points, with the calculated group mean (line) and standard error of the mean (SEM) or 95% confidence interval error bars for each group. As group sizes consisted of <12, two-sided, unpaired t tests using nonparametric statistics, Mann-Whitney U-tests were applied using an alpha of 0.05 for all analyses. Analyses were performed in GraphPad Prism (V7.0c, GraphPad Software).
Results
Meta-analysis of ACTN3 Arg577X Association Studies in Elite Sprint/Power Athletes Demonstrates a Consistent Homozygote Effect but a Heterogeneous 577RX Effect
Previous meta-analyses have assumed the association between human Arg577X genotype and elite international athlete status fits a dominant model26 and/or a recessive model27 but did not test whether data may better fit alternative models, such as an additive model. We therefore systematically searched relevant articles (Figure S1) and performed a meta-analysis using raw data from 12 independent studies that met a strict inclusion criterion (Table S1). This criterion aimed to minimize heterogeneity of the genetic effect by restricting analyses to similar-caliber athletes across studies who competed in similar events; in particular, by including study genotyping results comprised of >50%–100% international level (elite) athletes who had competed in an individual sprint/power event (Table S2). Elite endurance performance, such as the marathon, was not considered for meta-analysis purposes due to the absence of any consistent effect of the ACTN3 Arg577X variant.28
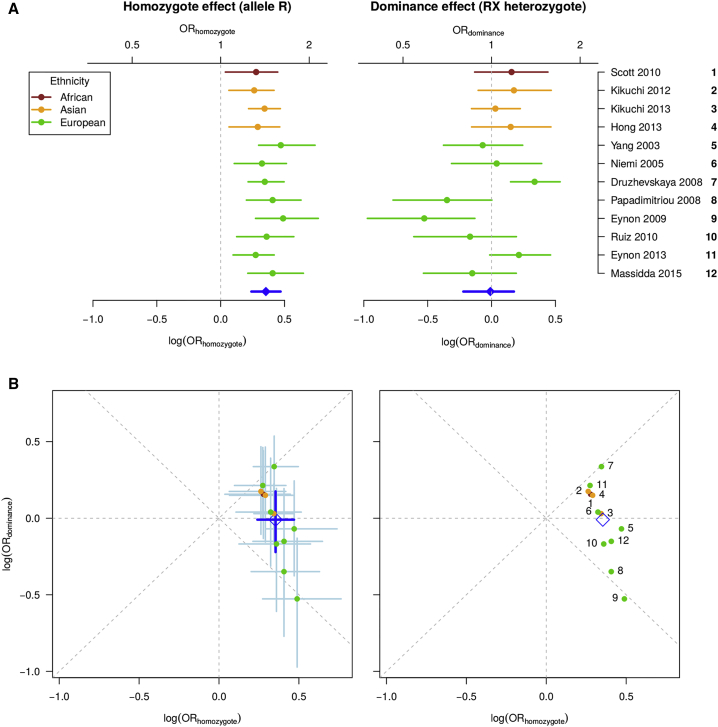
To analyze the effect of the ACTN3 genotype on sprint performance, we used a general genetic model for analyses of these data. This allowed us to assess the extent of evidence across the full range of simpler models (which includes dominant, additive, and recessive as special cases). This approach can be described as a Bayesian random effects meta-analysis and decomposes the association into two parts: (1) a “homozygote” component, which models the per-allele effect when comparing the two homozygotes (analogous to the “additive” component in a pure additive model); and (2) a “dominance” component, which models the extent to which the effect of the heterozygotes deviates from a purely additive (or dosage) effect. In particular, this second component can take values consistent with the overall model being dominant, recessive, or additive (and all models in between, see Figure S2 for visualization).
We found there was evidence for a consistent homozygote effect (overall per-allele odds ratio [OR] = 1.4, 95% CI 1.3–1.6; between-study SD τ = 0.12, 95% CI 0.01–0.31) (Figure 1). However, there was substantial heterogeneity in the dominance effect (overall OR = 0.98, 95% CI 0.73–1.3; between-study SD τ = 0.40, 95% CI 0.18–0.72). The variability was large, to the extent that some of the individual studies were consistent with a dominant model29 and some a recessive model30 (Figure S3), and overall, they were centered on an additive model (i.e., dominance OR of ≈ 1). Comparisons of both the homozygote and dominance effects against potential explanatory variables (ancestry, sex) did not show any clear trends (Figure S4) that were worth pursuing with formal heterogeneity measures (Supplemental Material and Methods). Overall, the meta-analysis showed that the difference between the homozygotes was consistent across these studies but no single genetic model can consistently explain the association between ACTN3 Arg577X and elite sprint/power performance due to the effect of heterozygotes being highly variable.
Figure 1.
Meta-analysis of the ACTN3 Arg577X Genotype Effect on Elite Power Performance
(A) Forrest plots with estimates of the posterior median and 90% credible interval of the odds ratios (OR) for each study are shown to demonstrate both the homozygote and dominance effects from the general meta-analysis model. Studies are grouped based on the ancestry of their participants, according to the key provided, and overall estimates are shown in blue.
(B) The same OR estimates are plotted jointly, with homozygote and dominance effects on the x and y axes, respectively. Diagonal gray lines correspond to dominant and recessive models. The point and interval estimates are identical to those in (A) and are numbered by study according to the key provided.
Overexpression of α-Actinin-3 Results in Reciprocal Downregulation of α-actinin-2 and Is Detrimental to WT Muscle Function
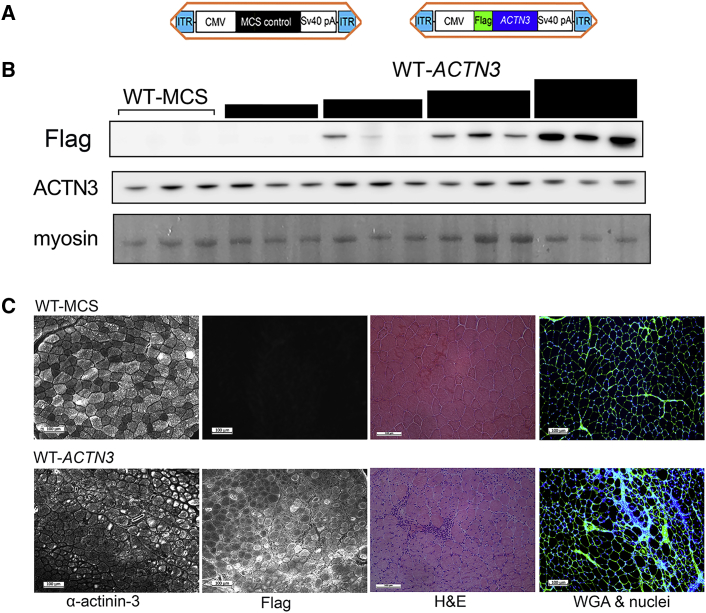
Given the ACTN3 577RR genotype-positive association with elite sprint/power performance, irrespective of the genotype model, we hypothesized that overexpression of ACTN3 in WT mice (“doping”) may still enhance muscle force generation and modify fatigue resistance. We have previously used rAAV-CMV-ACTN3 to induce α-actinin-3 overexpression in mouse muscles by intramuscular injections16 (Figure 2A); here we assess its effect on force and fatigue response at 5–6 weeks after injection. We examined the rAAV-CMV-ACTN3 vector expression at various doses (5E8, 1E9, 1E10, 1E11 vg) in WT muscles (WT-ACTN3) compared to muscles injected with empty vector (WT-MCS) (Figure 2B). Flag expression was highest at 1E11 vg, but there was no appreciable increases in α-actinin-3 expression at any dose examined.
Figure 2.
High Doses of rAAV-ACTN3 Cause Muscle Damage
(A) Schematics of the rAAV6 vector constructs. Expression of FLAG-tagged human ACTN3, or empty vector (MCS control), is driven by a CMV promoter, flanked by a SV-40 poly(A) tail, and capped by short inverted terminal repeats (ITRs).
(B) rAAV-ACTN3 vector expression, as shown by FLAG expression, increase with dose (5E8, 1E9, 1E10, and 1E11 vg) in WT (n = 3) muscles, indicative of efficient product expression. However, total ACTN3 protein did not increase with dose.
(C) Six weeks after high-dose injection (1E11 vg rAAV-ACTN3) into the WT TA muscles, cross-section shows positive flag staining accompanied by substantial centralized nuclei and cellular infiltration (H&E) and fibrosis (wheat germ agglutinin [WGA] staining).
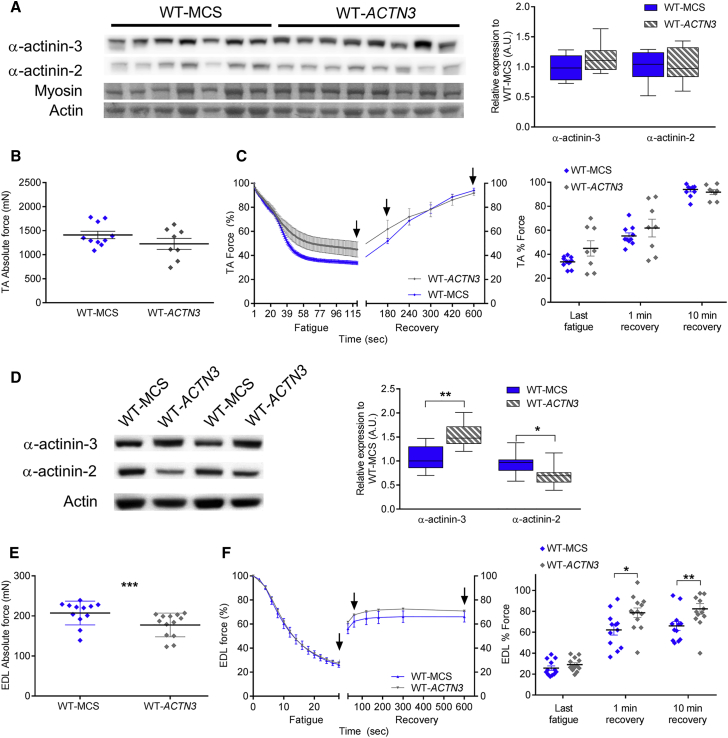
We further assessed the effect of α-actinin-3 overexpression in WT TA and EDL muscles at 1E11 vg. At this dose, ubiquitous rAAV-ACTN3 expression (as shown by Flag staining) caused wide-spread muscle damage, with increased centralized nuclei and inflammatory infiltrates relative to WT-MCS (Figure 2C). Staining with wheat germ agglutinin31 demonstrated increased fibrosis in WT-ACTN3 TA muscles. Desmin, an intermediate filament protein associated with muscle remodeling, was also upregulated in WT-ACTN3 TA muscles (Figure S5C). There was no overall increase in α-actinin-3 expression (Figure 3A), and no difference between WT-MCS and WT-ACTN3 TA muscles in maximal force generation and response to fatigue (Figures 3B and 3C).
Figure 3.
High Doses of rAAV-ACTN3 Intramuscular Injection (1E11 vg) Did Not Enhance Muscle Force in WT-ACTN3 Muscles
(A) α-Atinin-3 levels varied across injections in WT-ACTN3 TA (n = 8) compared to WT-MCS (n = 8), with no mean changes in α-actinin-3 or 2.
(B) No change in WT-ACTN3 TA force (1,227 ± 225 mN, n = 8) compared to WT-MCS (1,412 ± 152 mN, n = 10) 5 weeks after injection.
(C) Fatigue response and force recovery at 1 and 10 min (arrows) was also not different.
(D) α-Actinin-3 was increased in WT-ACTN3 EDL muscles compared to WT-MCS at 1E11 vg while α-actinin-2 is decreased.
(E) EDL force was lower in WT-ACTN3 (177 ± 16 mN, n = 13) compared to WT-MCS (207 ± 17 mN, n = 23) (p = 0.001).
(F) WT-ACTN3 EDL muscles showed improved recovery at 1 min and 10 min after fatigue compared to WT-MCS.
Boxplots represent the median, whiskers; min-max. Mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 Mann Whitney U test.
In contrast to TA muscles, the level of α-actinin-3 was ∼50% greater in WT-ACTN3 EDL muscles relative to WT-MCS EDL controls (p = 0.002). The increase in α-actinin-3 was accompanied by ∼30% downregulation of endogenous α-actinin-2, suggesting that expression of sarcomeric α-actinins (encoded by ACTN2 and ACTN3) is reciprocally regulated (p = 0.003) (Figure 3D). Despite increased α-actinin-3 expression, the maximal force in the WT-ACTN3 EDL was ∼15% lower than WT-MCS muscle (p < 0.001) (Figure 3E). In response to fatigue stimulation, WT-ACTN3 EDL muscles showed improved recovery from fatigue compared to the WT-MCS (p < 0.01) (Figure 3F). Overall, these results indicate that adult mammalian skeletal muscle fibers are sensitive to the levels of Actn3 expression and that supraphysiological overexpression of α-actinin-3 in mature skeletal muscle damages subcellular structure and is detrimental to muscle’s force-generating capacity.
Postnatal Replacement of α-Actinin-3 in Actn3 KO Muscles Does Not Rescue Force or Enhance Muscle Mass or Fast Fiber Size
Since supraphysiological overexpression of α-actinin-3 in WT muscles is damaging to skeletal muscle and detrimental to muscle force generation, we explored whether post-natal replacement of α-actinin-3 expression in Actn3 KO mice to WT levels could instead “rescue” the muscle force deficit in Actn3 KO muscles and increase susceptibility to fatigue to levels similar to that of WT muscles. We again performed a dose escalation assay with rAAV-CMV-ACTN3 (Figure S6) and determined that a dose of 1E9 vg was sufficient to replace α-actinin-3 expression in KO muscles to WT levels 6 weeks after injection.
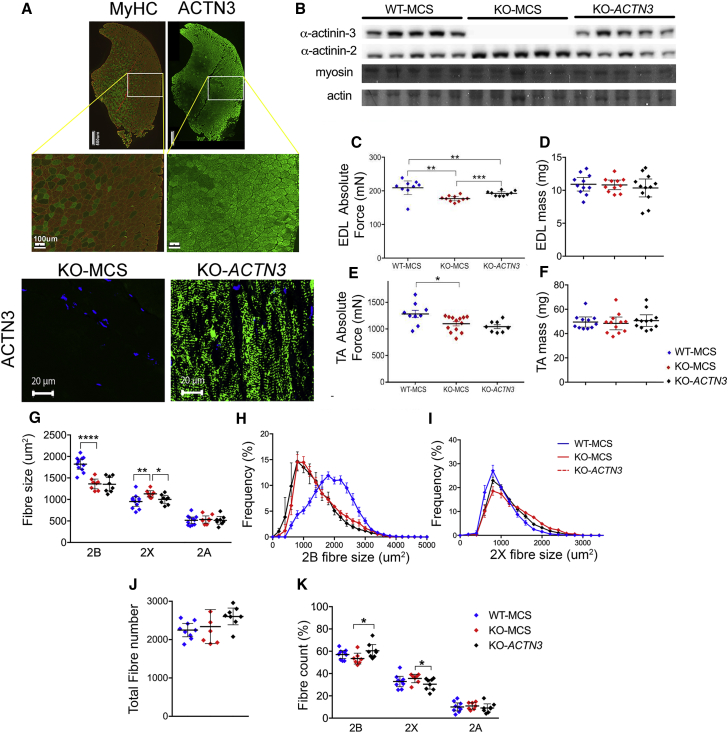
Despite ubiquitous expression of α-actinin-3 in all muscle fiber types, localization to the Z-disk (Figure 4A), and reciprocal downregulation of α-actinin-2 similar to WT-MCS muscles (Figure 4B), KO-ACTN3 TA (Figures 4C and 4D) and EDL (Figures 4E and 4F) muscles failed to reach the maximal forces elicited from WT-MCS muscles (Figures 4C and 4E). There was a small increase (5%) in absolute force in KO-ACTN3 EDL muscles relative to KO-MCS, but WT-MCS EDL absolute force was still 9% greater (p < 0.001) (Figure 4C). Similarly, KO-ACTN3 TA muscles demonstrated no improvement in muscle strength relative to KO-MCS, maintaining 18% lower absolute force compared to WT-MCS TA muscles (p < 0.05) (Figure 4E). There was also no effect on muscle mass in KO-ACTN3 EDL (Figure 4D) and TA (Figure 4F) muscles relative to KO-MCS. Mean fast 2B fiber size of KO-ACTN3 TA muscles was similar to KO-MCS and 20% smaller relative to WT-MCS (p < 0.0001) (Figures 4G and 4H). However, KO-ACTN3 2X fibers demonstrated a small reduction in size relative to KO-MCS (KO-ACTN3 1,000 ± 78 μm2, KO-MCS 1,037 ± 62 μm2 versus WT-MCS 951 ± 97 μm2) (p = 0.02) (Figure 4G), likely caused by increased proportions of smaller 2X fibers (Figure 4I). There was no change in total fiber count between groups (Figure 4J), but KO-ACTN3 muscles showed a small (6%) increase in 2B fiber proportions (p = 0.03) and a corresponding decrease in 2X fiber proportions relative to KO-MCS (p = 0.03) (Figure 4K).
Figure 4.
Postnatal Replacement of α-Actinin-3 in Actn3 KO Muscles Does Not Enhance Force, Muscle Mass, or Fast Fiber Size
(A) Intramuscular delivery of ACTN3 resulted in robust expression of α-actinin-3 in all fiber types. KO-ACTN3 TA muscle is immuno-labeled for myosin heavy chain 2B (red) and 2A (green) to denote fiber types. A longitudinally cut KO-ACTN3 muscle section stained for α-actinin-3 antibody (green), nuclei (blue) demonstrates localization to the Z-disk.
(B) KO-ACTN3 muscles show α-actinin-3 protein expression and reciprocal downregulation of α-actinin-2. Myosin and actin are loading controls.
(C) EDL force was increased in KO-ACTN3 (192 ± 5 mN) versus KO-MCS (177 ± 5 mN) but was lower than WT-MCS (210 ± 17 mN).
(D) EDL mass was not different between groups.
(E) KO-ACTN3 TA force is unchanged (1,044 ± 72 mN) compared to KO-MCS (1,099 ± 78 mN) and 18% lower than WT-MCS.
(F) TA mass.
(G–I) Mean 2B fiber size of KO-MCS and KO-ACTN3 TA muscles was not different, but 2X fiber size was reduced in KO-ACTN3 compared to KO-MCS, likely due to increased numbers of smaller 2X fibers.
(J) Total fiber number.
(K) KO-ACTN3 TA muscles have increased 2B and decreased 2X fiber proportions relative to KO-MCS.
N = 7–13 for all groups. Mean ± 95%CI. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001 Mann Whitney U.
Postnatal α-Actinin-3 Replacement in Actn3 KO Muscles Does Not Alter Calcineurin Activity or Expression of Other Z-disk Proteins
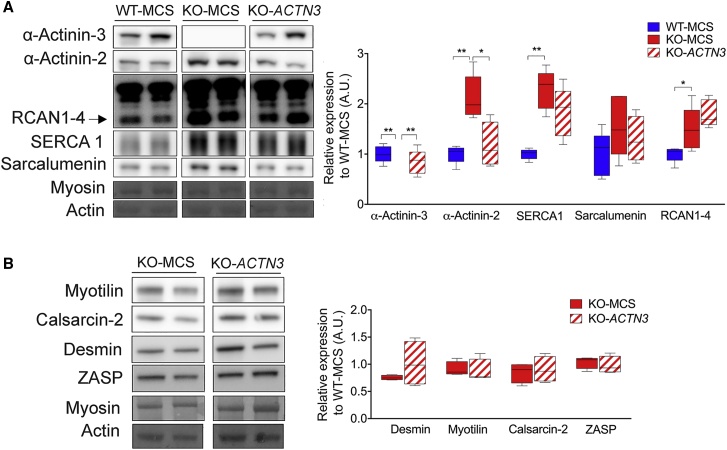
We further assessed RCAN1-4 expression as a marker of calcineurin activity to determine whether this could explain the shift in fiber type toward a fast-glycolytic phenotype. Calcineurin activity is calcium/calmodulin dependent and is inhibited by binding with calsarcin-2.32 However, in the absence of α-actinin-3, α-actinin-2 is upregulated and competes with calcineurin for binding with calsarcin-2, resulting in release of calcineurin from the inhibitory effect of calsarcin-2 and elevating calcineurin activity.16 We assessed RCAN1-4 expression by western blot, which is a downstream marker of calcineurin activity. Interestingly, despite a 28% downregulation of α-actinin-2 in KO-ACTN3 muscles relative to KO-MCS, RCAN1-4 expression was not significantly altered in TA muscles (Figure 5A). Calsarcin-2 (Figure 5B) and calcium signaling proteins SERCA1 and sarcalumenin (Figure 5A) were also not different between KO-ACTN3 and KO-MCS muscles.
Figure 5.
Postnatal α-Actinin-3 Replacement in Actn3 KO Muscles Did Not Alter Calcineurin Activity or Expression of Other Z-disk Proteins
(A) Calcineurin activity (as shown by RCAN1-4 expression) was unaltered in KO-ACTN3 muscles, despite reciprocal downregulation of α-actinin-2. Similarly, calcium handling proteins SERCA1 and sarcalumenin were unchanged in KO-ACTN3 muscles relative to KO-MCS.
(B) Expression of the Z-disk proteins. Myotilin, ZASP, calsarcin-2, and the remodeling protein, Desmin, was not altered in KO-ACTN3 muscles.
Mean ± 95%CI, N = 7–13 for all groups. Boxplots represent the median, whiskers; min-max. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Mann Whitney U.
Other Z-disk proteins, ZASP and myotilin and the intermediate filament protein desmin, upregulated in Actn3 KO muscles compared to WT,33 also remained unchanged in KO-ACTN3 muscles relative to KO-MCS (Figure 5B).
Postnatal Replacement of α-Actinin-3 in Actn3 KO Muscles Increases Muscle Fatigability and Reduces Force Recovery after Fatigue
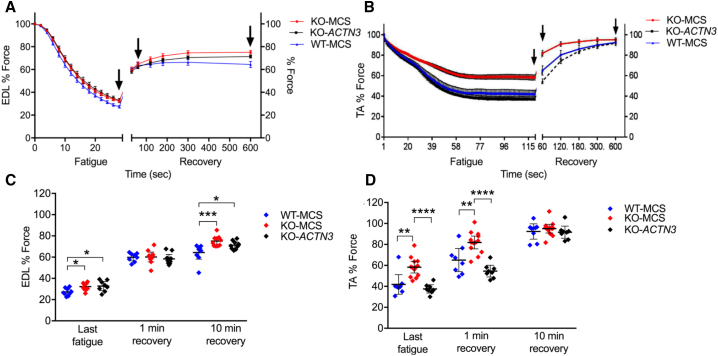
Given the baseline effects of α-actinin-3 deficiency in mice leading to enhanced recovery from fatigue,18 we recorded force recovery (super maximally stimulated) in WT-MCS, KO-MCS, and KO-ACTN3 muscles to determine whether overexpression of α-actinin-3 altered fatigue susceptibility. KO-ACTN3 muscles demonstrated alterations in fatigue resistance and force recovery relative to KO controls (Figure 6). KO-ACTN3 EDL muscles showed a trend for reduced force recovery compared to KO-MCS EDL muscle, but this did not reach statistical significance, with force recovery remaining higher relative to WT-MCS (p < 0.05) (Figures 6A and 6C). KO-ACTN3 TA muscles showed a fatigue profile that was more similar to WT-MCS TA muscles, with significantly higher fatigue and slower force recovery relative to KO-MCS TA muscles 1 min after fatigue (p < 0.0001) (Figures 6B and 6D). These results indicate that although post-natal restoration of α-actinin-3 in Actn3 KO TA muscles is unable to rescue force, there is a strong detrimental effect on the ability to recover from muscle fatigue.
Figure 6.
Postnatal Replacement of α-Actinin-3 in Actn3 KO Muscles Increases Muscle Fatigability and Reduces Force Recovery after Fatigue
(A and B) Force is reduced with repeated muscle stimulation to fatigue, and force recovery is assessed at 1, 2, 3, 5, and 10 min post-stimulation in EDL (A) and TA (B) muscles (mean ± SEM). Arrows indicate time points of cohort comparisons in the corresponding column graphs.
(C) KO-ACTN3 EDL demonstrated reduced force recovery at 10 min compared to the KO-MCS.
(D) KO-ACTN3 TA fatigued more rapidly than KO-MCS and were slower to recover after 1 min, similar to the WT-MCS.
Mean ± 95% CI ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Mann Whitney U, N = 6–13 for all groups.
Post-natal Replacement of α-Actinin-3 in Actn3 KO Muscle Results in a Shift to an Anaerobic Metabolic Profile
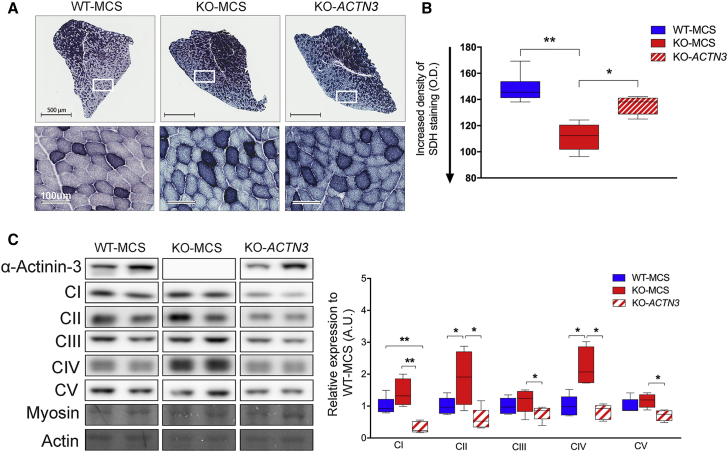
To determine the cause for the changes in muscle fatigue with post-natal expression of α-actinin-3 in KO muscle, we investigated the effect of skeletal muscle metabolism in KO-ACTN3 TA muscles. Succinate dehydrogenase activity (SDH; complex II) was assessed semiquantitatively by histochemical reaction staining on muscle cross-sections (Figures 7A and 7B). Cross-sectional areas positive for rAAV-ACTN3 were analyzed (n = 6 per group). These primarily consisted of 2B/2X fibers (the TA is 60% 2B/2X, Figure 4K) and demonstrated that KO-ACTN3 muscles stained less intensely for SDH than the KO-MCS control but remained significantly different compared to WT-MCS, suggesting partial shift of the muscle oxidative metabolism. We further assessed all five mitochondrial complexes in the TA by western blot (Figures 7C and S8) (n = 5 per group). At baseline, complexes II and IV were increased in KO-MCS muscles compared to WT-MCS; these correspond to SDH and the cytochrome-c oxidase subunit I, respectively. We have previously shown that NADH, SDH, and CIV activity is upregulated in KO muscles.34 Interestingly, replacement of α-actinin-3 in KO TA muscles resulted in significant decreases in all complexes (I, II, III, IV, and V) relative to KO-MCS, to be equal or below WT-MCS control levels. These results suggest that α-actinin-3 influences mitochondrial activity via specific effects on complexes on the electron transport chain.
Figure 7.
α-Actinin-3 Replacement in KO TA Muscles Reduces Mitochondrial Oxidative Metabolism
(A) Succinate dehydrogenase activity staining in WT-MCS, KO-MCS, and KO-ACTN3 TA muscle.
(B) Intensity of positive blue staining is reduced in KO-ACTN3 relative to KO-MCS (indicative of lower SDH activity) but remains different compared to the WT-MCS, N = 6 all groups.
(C) KO-ACTN3 TA muscle demonstrated decreases in mitochondrial complexes I, II, III, IV, and V to WT levels in response to α-actinin-3 expression. N = 5 for all groups.
Boxplots represent the median, whiskers; min-max. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
α-Actinin-3 Overexpression (WT-ACTN3) Damages Skeletal Muscle: The Case against Gene Doping
Results from our meta-analysis confirmed that complete absence of α-actinin-3 (ACTN3 577XX) is detrimental for elite sprint performance compared to 577RR homozygotes. Heterozygotes were intermediate (RR > RX > XX) but there was substantial variation across studies. Larger sample sizes, expression levels, and genome-wide testing in elite-international cohorts may help understand these population-specific complexities noted here in our general model meta-analysis. The consistent positive effect associated with ACTN3 577RR homozygotes in human sprint/power performance supported the hypothesis that rAAV gene delivery of ACTN3 would enhance muscle performance. Here we have tested multiple doses of rAAV-ACTN3 in our wild-type (WT) mouse line, in two different muscles over 6 weeks, and demonstrated that overexpression of α-actinin-3 in mature skeletal muscle had no immediate effect on force. Furthermore, at high dose levels, ACTN3 overexpression was detrimental to muscle cytoarchitecture and function, with muscles showing reduced force generation. Force generation and recovery after fatigue was incrementally altered in WT mice with rAAV-ACTN3 injected into the EDL muscles but this was not replicated in TA muscles.
The presence of fibrosis and centralized nuclei in the WT muscles injected with rAAV-ACTN3 indicates that supraphysiological ACTN3 overexpression for 6 weeks triggered recurrent muscle damage and remodelling. The responses in the EDL may be an artifact related to reduced work performed by damaged/regenerating muscles.35 This is unlikely to be due to the presentation of foreign antigens as a result of species differences in mouse and human α-actinin-3, as they share 97% identity. Furthermore, expression of ACTN3 in Actn3 knock-out (KO) muscles, which were devoid of α-actinin-3 from birth, and low doses (5E8–1E10) of rAAV-ACTN3 in WT mice did not result in any consistent muscle damage. One explanation for the presence of recurring muscle damage may be associated with the level of α-actinin overexpression and the length of vector expression. We note that no consistent muscle damage was observed in WT muscles injected with ≤1E10 vector genomes (vg) of rAAV-ACTN3 for 6 weeks, or in muscles injected with 1E11 vg of rAAV-ACTN3 for 1 and 2 weeks (Figures S5 and S7). This suggests that muscle fibers can incorporate additional sarcomeric α-actinin up to a certain threshold, consistent with normal 2- to 3-fold variation in Z-line layers between fiber types.36, 37 Excessive α-actinin-3 expression, however, whether this is dispersed across the whole muscle or localized to a certain region due to variation in delivery, has a “toxic” effect on skeletal muscles. This sensitive balance has previously been shown with other contractile proteins.38, 39, 40 For example, moderate overexpression of titin (the Z-band region) localized normally at the Z-line in contracting myofibrils and normal function was observed. However, high levels caused myofibril disassembly as the fragments accumulated.41 High expression of desmin has also been found to form a reticular intermediate filament meshwork and non-filamentous aggregates.42
In WT quadriceps muscles at baseline (with a large proportion of fast fibers 2B fibers), sarcomeric α-actinin expression in skeletal muscle is maintained at a ratio of 1:1 for α-actinin-3 and α-actinin-2.32 Therefore, the muscle damage observed in WT-ACTN3 may be associated with loss of the optimal ratio of relative expression of the two sarcomeric α-actinins. Overexpression of α-actinin-3 in WT muscles was not accompanied by an equivalent level of α-actinin-2 downregulation, regardless of rAAV dose (Figure 2). It is possible that high, non-toxic, levels of α-actinin-3 may be achieved only if there is parallel α-actinin-2 knockdown that we did not investigate. We note that maintenance of α-actinin-2 may reflect an essential role in normal muscle function. Knockdown of α-actinin-2 in zebrafish results in sarcomeric disorganization in skeletal muscles and is not compensated by increased endogenous α-actinin-3.43 This contrasts with α-actinin-3 deficiency, which does demonstrate partial compensation by endogenous upregulation of ACTN2 in muscles of mice18, 21 and humans (controlled for fiber type)44 and results in normal, highly organized, and functional sarcomeres. Moreover, in zebrafish, replacement with actn3a, actn3b, or human ACTN3 fails to ameliorate the developmental defects associated with α-actinin-2 deficiency,43 highlighting the functional differences between α-actinin-2 and α-actinin-3.
α-Actinin-3 Rescue (KO-ACTN3) Does Not Restore Muscle Mass and Strength to Wild-Type Levels and Is Unlikely to Confer a Therapeutic Benefit in Muscle Wasting Diseases
Contrary to expectations, postnatal replacement of α-actinin-3 in Actn3 KO muscles failed to restore force production, muscle mass, and fast fiber size to WT levels, despite 6 weeks of gene expression and reintroduction of α-actinin-3 to ∼64% of WT levels (Figure 4). At a dose of 1E9 vg, α-actinin-2 was also reciprocally downregulated in KO-ACTN3 muscles by approximately equal proportions relative to KO controls. This mimics the normal developmental regulation of total α-actinin levels in the WT muscles (where α-actinin-3 is detectable from 2 weeks of age and is associated with downregulation of α-actinin-214) and the dose-dependent upregulation of α-actinin-2 in the Actn3 KO and HET mice.18, 21 However, the reincorporation of α-actinin-3 in Actn3 KO muscle Z-disks was not accompanied by a change in Z-disk structural protein composition. Despite a ∼56% reduction of α-actinin-2, myotilin, calsarcin-2, and ZASP levels were unchanged in KO-ACTN3 muscles, whereas we would have predicted a decrease in levels (Figure 5). Contrary to expectations, RCAN1-4 expression was also unchanged. We have previously shown that upregulation of α-actinin-2 results in increased activity of calcineurin activity through its release from the inhibitory effects of calsarcin-2,16 so a reduction in RCAN1-4 was expected. It is possible that the reduction in α-actinin-2 in KO-ACTN3 muscles was insufficient to significantly influence calcineurin activity, since these muscles still express 44% higher levels of α-actinin-2 compared to WT. This is supported by the presence of only minor (6%–10%) fiber type shift toward fast 2B (Figure 4K). The lack of change in RCAN1-4 and Z-disk structural protein expression in KO-ACTN3 muscles is also consistent with a lack of muscle damage and regeneration.
The absence of any meaningful effect on muscle mass and force with postnatal α-actinin-3 replacement in Actn3 KO muscles indicates that this model is not recapitulating muscle phenotypes of Actn3 WT and heterozygous mice, which express α-actinin-3 from birth. These results also suggest that the pivotal effects of α-actinin-3 on muscle traits begin during skeletal muscle development. α-Actinin-3 is detectable in mouse skeletal muscle at E14.5,1 a time point that coincides with the end of primary myogenesis, during which myotubes lengthen as new nuclei fuse with primary myotubes.45 Therefore, α-actinin-3 may, in fact, play a role in regulating fiber size and muscle growth during embryonic muscle development. Calcineurin signaling is also known to govern the formation, growth, and maturation of the musculature through specific, temporal expression of NFATc246 and NFATc3.47 In the absence of α-actinin-3, calcineurin activity is elevated in mature muscles.16 This raises the possibility that ACTN3 genotype influences fiber size and muscle mass via calcineurin signaling during muscle development.
Despite the absence of an effect on muscle mass and strength, replacement of α-actinin-3 in Actn3 KO muscles did significantly alter skeletal muscle response to fatigue. KO-ACTN3 muscles showed a fatigue profile that was more similar to WT muscles, producing 20% lower maximum relative force compared to KO-MCS after 2 min of repeated electrical stimulation (Figure 6D). The shift toward increased fatigability was expected, since α-actinin-3 deficiency is associated with enhanced force recovery after fatigue, increased endurance capacity, and response to exercise training.16, 18 Analysis of mitochondrial enzyme activity showed that increased fatigability of KO-ACTN3 muscles was associated with marked decreases in expression of mitochondrial complexes I-V, which include NADH dehydrogenase, SDH, and the ubiquinol-cytochrome c reductase complex (Figures 7 and S8). Intriguingly, the expression of some of these complexes in KO-ACTN3 muscles are on par with levels found in WT muscles, suggesting that the primary role of α-actinin-3 in skeletal muscle is the regulation of muscle metabolism, specifically, mitochondrial enzyme activity. Moreover, the lack of change in other measured phenotypes in KO-ACTN3 muscles suggest that the effect of α-actinin-3 on oxidative metabolism in skeletal muscle occurs prior to, or independent of, its effects on Z-disk protein composition, fast fiber size, and calcium/calcineurin signaling.16, 18
Our present study thus confirms that a primary function of α-actinin-3 in skeletal muscle is to regulate muscle metabolism, and this precedes or occurs independently of its other functions in skeletal muscle. This supports the shift in muscle metabolism as a key driver for the positive selection of the ACTN3 577X allele during recent human evolution and the functional specialization of ACTN2 and ACTN3 genes following divergence.34, 48 We suggest, however, that these metabolic effects may be specific to muscle. The prevalence of metabolic disease is highly influenced by genetic factors49 and while a number of studies have investigated ACTN3 Arg577X as a modifier of these complex traits,17, 50, 51 no ACTN3-specific loci effect has been identified genome-wide. Further expression studies focused on the skeletal adaptation effects, such as denervation or exercise training seen in both human and mouse studies,16, 52, 53, 54 may provide greater insight into the effect of ACTN3. Specifically, our results demonstrate that the metabolic impacts of ACTN3 may not be limited to the calcineurin pathway and other mechanisms may also be at play. In the context of gene-doping risks and gene therapy, our findings demonstrate the sensitive balance of sarcomeric α-actinin expression at the Z-line. Both in vivo rescue and overexpression of ACTN3 reveals insight into its metabolic role in skeletal muscle, but this is innately linked to the level of expression which can be easily disrupted causing detrimental functional effects, reminiscent of disease.
Acknowledgments
We thank Dr. Hongwei Qian (Baker Institute, Melbourne) for assistance with production of the recombinant AAV vectors used for these studies and Rachel E. Thomson (Baker Institute, Melbourne) and Sharon Cunningham (Children’s Medical Research Institute, Sydney) for rAAV advice. This project was funded in part by grants from the National Health and Medical Research Council of Australia (NHMRC) (1002033 and 1062500 to K.N.N.). P.G. is supported by a senior research fellowship (SRFA APP1046782, NHMRC) and J.T.S. and F.C.G. are supported by early career research fellowships (APP1036656, APP1109782, NHMRC). K.N.R. and F.X.Z.L. are supported by Australian Postgraduate awards (NHMRC). Both MCRI and the Baker Institute are recipients of the Victorian Government Operational Infrastructure support scheme. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Published: April 26, 2018
Footnotes
Supplemental Data include Supplemental Material and Methods, eight figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.009.
Supplemental Data
References
- 1.Mills M., Yang N., Weinberger R., Vander Woude D.L., Beggs A.H., Easteal S., North K. Differential expression of the actin-binding proteins, alpha-actinin-2 and -3, in different species: implications for the evolution of functional redundancy. Hum. Mol. Genet. 2001;10:1335–1346. doi: 10.1093/hmg/10.13.1335. [DOI] [PubMed] [Google Scholar]
- 2.North K.N., Yang N., Wattanasirichaigoon D., Mills M., Easteal S., Beggs A.H. A common nonsense mutation results in alpha-actinin-3 deficiency in the general population. Nat. Genet. 1999;21:353–354. doi: 10.1038/7675. [DOI] [PubMed] [Google Scholar]
- 3.Eynon N., Hanson E.D., Lucia A., Houweling P.J., Garton F., North K.N., Bishop D.J. Genes for elite power and sprint performance: ACTN3 leads the way. Sports Med. 2013;43:803–817. doi: 10.1007/s40279-013-0059-4. [DOI] [PubMed] [Google Scholar]
- 4.Garton F.C., North K.N. The effect of heterozygosity for the ACTN3 null allele on human muscle performance. Med. Sci. Sports Exerc. 2016;48:509–520. doi: 10.1249/MSS.0000000000000784. [DOI] [PubMed] [Google Scholar]
- 5.Willems S., Wright D., Day F., Trajanoska K., Joshi P., Morris J.A., Matteini A., Garton F.C., Grarup N., Oskolkov N. Large-scale GWAS identifies sixteen loci associated with hand grip strength and provides new insights into the biology of muscular fitness. Nat. Commun. 2017 doi: 10.1038/ncomms16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zempo H., Tanabe K., Murakami H., Iemitsu M., Maeda S., Kuno S. Age differences in the relation between ACTN3 R577X polymorphism and thigh-muscle cross-sectional area in women. Genet. Test. Mol. Biomarkers. 2011;15:639–643. doi: 10.1089/gtmb.2011.0005. [DOI] [PubMed] [Google Scholar]
- 7.Yang N., Schindeler A., McDonald M.M., Seto J.T., Houweling P.J., Lek M., Hogarth M., Morse A.R., Raftery J.M., Balasuriya D. α-Actinin-3 deficiency is associated with reduced bone mass in human and mouse. Bone. 2011;49:790–798. doi: 10.1016/j.bone.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Shang X., Li Z., Cao X., Xie C., Gu M., Chen P., Yang X., Cai J. The association between the ACTN3 R577X polymorphism and noncontact acute ankle sprains. J. Sports Sci. 2015;33:1775–1779. doi: 10.1080/02640414.2015.1012098. [DOI] [PubMed] [Google Scholar]
- 9.Judson R.N., Wackerhage H., Hughes A., Mavroeidi A., Barr R.J., Macdonald H.M., Ratkevicius A., Reid D.M., Hocking L.J. The functional ACTN3 577X variant increases the risk of falling in older females: results from two large independent cohort studies. J. Gerontol. A Biol. Sci. Med. Sci. 2011;66:130–135. doi: 10.1093/gerona/glq189. [DOI] [PubMed] [Google Scholar]
- 10.De Filippi P., Saeidi K., Ravaglia S., Dardis A., Angelini C., Mongini T., Morandi L., Moggio M., Di Muzio A., Filosto M. Genotype-phenotype correlation in Pompe disease, a step forward. Orphanet J. Rare Dis. 2014;9:102. doi: 10.1186/s13023-014-0102-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogarth M.W., Houweling P.J., Thomas K.C., Gordish-Dressman H., Bello L., Pegoraro E., Hoffman E.P., Head S.I., North K.N., Cooperative International Neuromuscular Research Group (CINRG) Evidence for ACTN3 as a genetic modifier of Duchenne muscular dystrophy. Nat. Commun. 2017;8:14143. doi: 10.1038/ncomms14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vincent B., De Bock K., Ramaekers M., Van den Eede E., Van Leemputte M., Hespel P., Thomis M.A. ACTN3 (R577X) genotype is associated with fiber type distribution. Physiol. Genomics. 2007;32:58–63. doi: 10.1152/physiolgenomics.00173.2007. [DOI] [PubMed] [Google Scholar]
- 13.Ahmetov I.I., Druzhevskaya A.M., Lyubaeva E.V., Popov D.V., Vinogradova O.L., Williams A.G. The dependence of preferred competitive racing distance on muscle fibre type composition and ACTN3 genotype in speed skaters. Exp. Physiol. 2011;96:1302–1310. doi: 10.1113/expphysiol.2011.060293. [DOI] [PubMed] [Google Scholar]
- 14.Quinlan K.G., Seto J.T., Turner N., Vandebrouck A., Floetenmeyer M., Macarthur D.G., Raftery J.M., Lek M., Yang N., Parton R.G. α-actinin-3 deficiency results in reduced glycogen phosphorylase activity and altered calcium handling in skeletal muscle. Hum. Mol. Genet. 2010;19:1335–1346. doi: 10.1093/hmg/ddq010. [DOI] [PubMed] [Google Scholar]
- 15.Norman B., Esbjörnsson M., Rundqvist H., Osterlund T., von Walden F., Tesch P.A. Strength, power, fiber types, and mRNA expression in trained men and women with different ACTN3 R577X genotypes. J. Appl. Physiol. 2009;106:959–965. doi: 10.1152/japplphysiol.91435.2008. [DOI] [PubMed] [Google Scholar]
- 16.Seto J.T., Quinlan K.G., Lek M., Zheng X.F., Garton F., MacArthur D.G., Hogarth M.W., Houweling P.J., Gregorevic P., Turner N. ACTN3 genotype influences muscle performance through the regulation of calcineurin signaling. J. Clin. Invest. 2013;123:4255–4263. doi: 10.1172/JCI67691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riedl I., Osler M.E., Benziane B., Chibalin A.V., Zierath J.R. Association of the ACTN3 R577X polymorphism with glucose tolerance and gene expression of sarcomeric proteins in human skeletal muscle. Physiol. Rep. 2015;3:3. doi: 10.14814/phy2.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacArthur D.G., Seto J.T., Chan S., Quinlan K.G., Raftery J.M., Turner N., Nicholson M.D., Kee A.J., Hardeman E.C., Gunning P.W. An Actn3 knockout mouse provides mechanistic insights into the association between alpha-actinin-3 deficiency and human athletic performance. Hum. Mol. Genet. 2008;17:1076–1086. doi: 10.1093/hmg/ddm380. [DOI] [PubMed] [Google Scholar]
- 19.Seto J.T., Chan S., Turner N., MacArthur D.G., Raftery J.M., Berman Y.D., Quinlan K.G., Cooney G.J., Head S., Yang N., North K.N. The effect of α-actinin-3 deficiency on muscle aging. Exp. Gerontol. 2011;46:292–302. doi: 10.1016/j.exger.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Garton F.C., Seto J.T., Quinlan K.G., Yang N., Houweling P.J., North K.N. α-Actinin-3 deficiency alters muscle adaptation in response to denervation and immobilization. Hum. Mol. Genet. 2014;23:1879–1893. doi: 10.1093/hmg/ddt580. [DOI] [PubMed] [Google Scholar]
- 21.Hogarth M.W., Garton F.C., Houweling P.J., Tukiainen T., Lek M., Macarthur D.G., Seto J.T., Quinlan K.G., Yang N., Head S.I., North K.N. Analysis of the ACTN3 heterozygous genotype suggests that α-actinin-3 controls sarcomeric composition and muscle function in a dose-dependent fashion. Hum. Mol. Genet. 2016;25:866–877. doi: 10.1093/hmg/ddv613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregorevic P., Blankinship M.J., Allen J.M., Crawford R.W., Meuse L., Miller D.G., Russell D.W., Chamberlain J.S. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat. Med. 2004;10:828–834. doi: 10.1038/nm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmeli E., Coleman R., Reznick A.Z. The biochemistry of aging muscle. Exp. Gerontol. 2002;37:477–489. doi: 10.1016/s0531-5565(01)00220-0. [DOI] [PubMed] [Google Scholar]
- 24.Chan S., Seto J.T., MacArthur D.G., Yang N., North K.N., Head S.I. A gene for speed: contractile properties of isolated whole EDL muscle from an α-actinin-3 knockout mouse. Am. J. Physiol. Cell Physiol. 2008;295:C897–C904. doi: 10.1152/ajpcell.00179.2008. [DOI] [PubMed] [Google Scholar]
- 25.Garton F., Seto J.T., North K.N., Yang N. Validation of an automated computational method for skeletal muscle fibre morphometry analysis. Neuromuscul. Disord. 2010;20:540–547. doi: 10.1016/j.nmd.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Alfred T., Ben-Shlomo Y., Cooper R., Hardy R., Cooper C., Deary I.J., Gunnell D., Harris S.E., Kumari M., Martin R.M., HALCyon study team ACTN3 genotype, athletic status, and life course physical capability: meta-analysis of the published literature and findings from nine studies. Hum. Mutat. 2011;32:1008–1018. doi: 10.1002/humu.21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma F., Yang Y., Li X., Zhou F., Gao C., Li M., Gao L. The association of sport performance with ACE and ACTN3 genetic polymorphisms: a systematic review and meta-analysis. PLoS ONE. 2013;8:e54685. doi: 10.1371/journal.pone.0054685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang N., Garton F., North K. alpha-actinin-3 and performance. Med. Sport Sci. 2009;54:88–101. doi: 10.1159/000235698. [DOI] [PubMed] [Google Scholar]
- 29.Druzhevskaya A.M., Ahmetov I.I., Astratenkova I.V., Rogozkin V.A. Association of the ACTN3 R577X polymorphism with power athlete status in Russians. Eur. J. Appl. Physiol. 2008;103:631–634. doi: 10.1007/s00421-008-0763-1. [DOI] [PubMed] [Google Scholar]
- 30.Papadimitriou I.D., Papadopoulos C., Kouvatsi A., Triantaphyllidis C. The ACTN3 gene in elite Greek track and field athletes. Int. J. Sports Med. 2008;29:352–355. doi: 10.1055/s-2007-965339. [DOI] [PubMed] [Google Scholar]
- 31.Emde B., Heinen A., Gödecke A., Bottermann K. Wheat germ agglutinin staining as a suitable method for detection and quantification of fibrosis in cardiac tissue after myocardial infarction. Eur. J. Histochem. 2014;58:2448. doi: 10.4081/ejh.2014.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frey N., Frank D., Lippl S., Kuhn C., Kögler H., Barrientos T., Rohr C., Will R., Müller O.J., Weiler H. Calsarcin-2 deficiency increases exercise capacity in mice through calcineurin/NFAT activation. J. Clin. Invest. 2008;118:3598–3608. doi: 10.1172/JCI36277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seto J.T., Lek M., Quinlan K.G., Houweling P.J., Zheng X.F., Garton F., MacArthur D.G., Raftery J.M., Garvey S.M., Hauser M.A. Deficiency of α-actinin-3 is associated with increased susceptibility to contraction-induced damage and skeletal muscle remodeling. Hum. Mol. Genet. 2011;20:2914–2927. doi: 10.1093/hmg/ddr196. [DOI] [PubMed] [Google Scholar]
- 34.MacArthur D.G., Seto J.T., Raftery J.M., Quinlan K.G., Huttley G.A., Hook J.W., Lemckert F.A., Kee A.J., Edwards M.R., Berman Y. Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. Nat. Genet. 2007;39:1261–1265. doi: 10.1038/ng2122. [DOI] [PubMed] [Google Scholar]
- 35.Bogdanis G.C. Effects of physical activity and inactivity on muscle fatigue. Front. Physiol. 2012;3:142. doi: 10.3389/fphys.2012.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgoyne T., Morris E.P., Luther P.K. Three-dimensional structure of vertebrate muscle Z-band: the small-square lattice Z-band in rat cardiac muscle. J. Mol. Biol. 2015;427:3527–3537. doi: 10.1016/j.jmb.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luther P.K. The vertebrate muscle Z-disc: sarcomere anchor for structure and signalling. J. Muscle Res. Cell Motil. 2009;30:171–185. doi: 10.1007/s10974-009-9189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joanne P., Chourbagi O., Hourdé C., Ferry A., Butler-Browne G., Vicart P., Dumonceaux J., Agbulut O. Viral-mediated expression of desmin mutants to create mouse models of myofibrillar myopathy. Skelet. Muscle. 2013;3:4. doi: 10.1186/2044-5040-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garvey S.M., Miller S.E., Claflin D.R., Faulkner J.A., Hauser M.A. Transgenic mice expressing the myotilin T57I mutation unite the pathology associated with LGMD1A and MFM. Hum. Mol. Genet. 2006;15:2348–2362. doi: 10.1093/hmg/ddl160. [DOI] [PubMed] [Google Scholar]
- 40.Rodríguez Fernández J.L., Geiger B., Salomon D., Ben-Ze’ev A. Overexpression of vinculin suppresses cell motility in BALB/c 3T3 cells. Cell Motil. Cytoskeleton. 1992;22:127–134. doi: 10.1002/cm.970220206. [DOI] [PubMed] [Google Scholar]
- 41.Turnacioglu K.K., Mittal B., Dabiri G.A., Sanger J.M., Sanger J.W. An N-terminal fragment of titin coupled to green fluorescent protein localizes to the Z-bands in living muscle cells: overexpression leads to myofibril disassembly. Mol. Biol. Cell. 1997;8:705–717. doi: 10.1091/mbc.8.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cary R.B., Klymkowsky M.W. Differential organization of desmin and vimentin in muscle is due to differences in their head domains. J. Cell Biol. 1994;126:445–456. doi: 10.1083/jcb.126.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta V., Discenza M., Guyon J.R., Kunkel L.M., Beggs A.H. α-Actinin-2 deficiency results in sarcomeric defects in zebrafish that cannot be rescued by α-actinin-3 revealing functional differences between sarcomeric isoforms. FASEB J. 2012;26:1892–1908. doi: 10.1096/fj.11-194548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norman B., Esbjörnsson M., Rundqvist H., Österlund T., Glenmark B., Jansson E. ACTN3 genotype and modulation of skeletal muscle response to exercise in human subjects. J. Appl. Physiol. 2014;116:1197–1203. doi: 10.1152/japplphysiol.00557.2013. [DOI] [PubMed] [Google Scholar]
- 45.Ontell M. Neonatal muscle: an electron microscopic study. Anat. Rec. 1977;189:669–690. doi: 10.1002/ar.1091890410. [DOI] [PubMed] [Google Scholar]
- 46.Horsley V., Friday B.B., Matteson S., Kegley K.M., Gephart J., Pavlath G.K. Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J. Cell Biol. 2001;153:329–338. doi: 10.1083/jcb.153.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kegley K.M., Gephart J., Warren G.L., Pavlath G.K. Altered primary myogenesis in NFATC3(-/-) mice leads to decreased muscle size in the adult. Dev. Biol. 2001;232:115–126. doi: 10.1006/dbio.2001.0179. [DOI] [PubMed] [Google Scholar]
- 48.Lek M., Quinlan K.G., North K.N. The evolution of skeletal muscle performance: gene duplication and divergence of human sarcomeric alpha-actinins. BioEssays. 2010;32:17–25. doi: 10.1002/bies.200900110. [DOI] [PubMed] [Google Scholar]
- 49.Polderman T.J., Benyamin B., de Leeuw C.A., Sullivan P.F., van Bochoven A., Visscher P.M., Posthuma D. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet. 2015;47:702–709. doi: 10.1038/ng.3285. [DOI] [PubMed] [Google Scholar]
- 50.Bernardez-Pereira S., Santos P.C., Krieger J.E., Mansur A.J., Pereira A.C. ACTN3 R577X polymorphism and long-term survival in patients with chronic heart failure. BMC Cardiovasc. Disord. 2014;14:90. doi: 10.1186/1471-2261-14-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim K., Ahn N., Park J., Koh J., Jung S., Kim S., Moon S. Association of angiotensin-converting enzyme I/D and α-actinin-3 R577X genotypes with metabolic syndrome risk factors in Korean children. Obes. Res. Clin. Pract. 2016;10(Suppl 1):S125–S132. doi: 10.1016/j.orcp.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Clarkson P.M., Devaney J.M., Gordish-Dressman H., Thompson P.D., Hubal M.J., Urso M., Price T.B., Angelopoulos T.J., Gordon P.M., Moyna N.M. ACTN3 genotype is associated with increases in muscle strength in response to resistance training in women. J. Appl. Physiol. 2005;99:154–163. doi: 10.1152/japplphysiol.01139.2004. [DOI] [PubMed] [Google Scholar]
- 53.Delmonico M.J., Kostek M.C., Doldo N.A., Hand B.D., Walsh S., Conway J.M., Carignan C.R., Roth S.M., Hurley B.F. Alpha-actinin-3 (ACTN3) R577X polymorphism influences knee extensor peak power response to strength training in older men and women. J. Gerontol. A Biol. Sci. Med. Sci. 2007;62:206–212. doi: 10.1093/gerona/62.2.206. [DOI] [PubMed] [Google Scholar]
- 54.Gentil P., Pereira R.W., Leite T.K., Bottaro M. ACTN3 R577X polymorphism and neuromuscular response to resistance training. J. Sports Sci. Med. 2011;10:393–399. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.