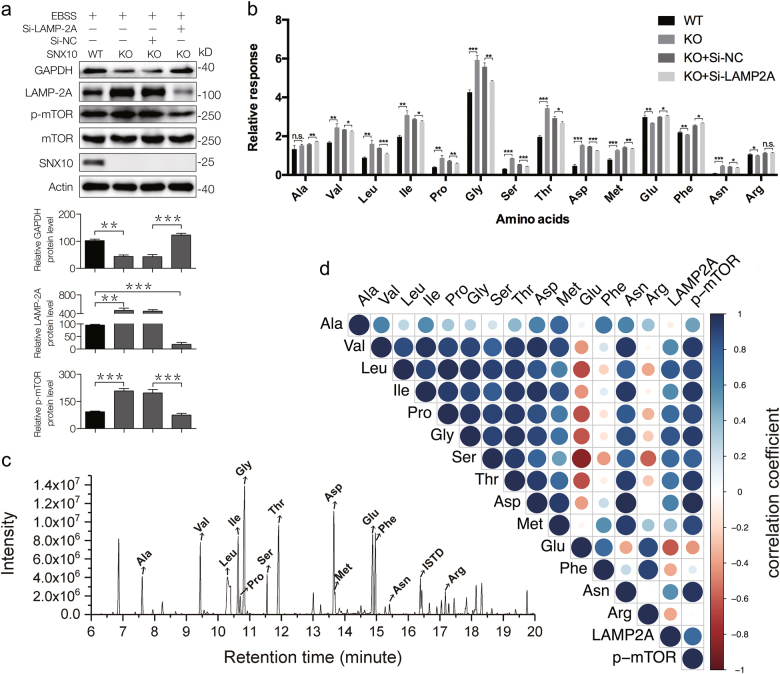
Fig. 6. Targeted metabolomics analysis of amino acids using GC-MS validated that SNX10 deficiency enhanced CMA process leading to the accumulation of specific amino acids that can activate mTORC1 in CRC cells.
a WT, SNX10 KO HCT116 cells, and SNX10 KO HCT116 cells transfected with nontargeting siRNA (Si-NC) or LAMP-2A siRNA (Si-LAMP-2A) were treated with EBSS for 24 h, the total proteins were isolated and immunobloted with indicated antibodies. The bands of GAPDH, LAMP-2A, and p-mTOR were quantified using Quantity One software. b Relative levels of amino acids in WT, SNX10 KO and transfected with nontargeting siRNA (Si-NC) or LAMP-2A siRNA (Si-LAMP-2A) HCT116 cells. c Represented chromatogram of targeted metabolomics analysis of amino acids using GC-MS. d Pearson correlation analysis of amino-acid cellular concentrations with LAMP-2A and p-mTOR levels, blue dots represent positive correlation and red dots represent negative correlation, the deeper the color of the dots the greater the radius and the stronger the correlation. Data are derived from at least three independent experiments and represented as means ± SEM in the bar graphs. Not significant (n.s.), *P < 0.05, **P < 0.01, and ***P < 0.001