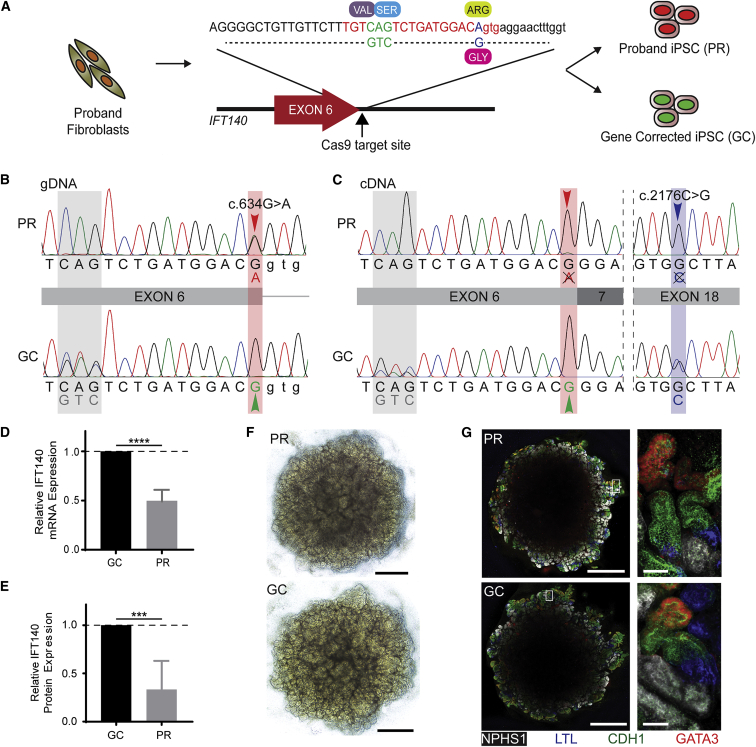
Figure 2.
Gene Correction, Transcript Analysis, and Differentiation to Kidney Organoid
(A) The gene-correction template for the c.634G>A variant contains the corrected 1 bp substitution (blue) and a synonymous downstream 3 bp substitution (green) for ease of identification by Sanger sequencing. Successfully reprogrammed clones were screened for unsuccessful (red nuclei, PR) or successful (green nuclei, GC) gene correction.
(B) DNA sequencing demonstrates the c.634G>A variant in the PR clone (red arrowhead) and successful correction of this variant in the GC clone (green arrowhead), as well as the synonymous 3 bp change in this allele (gray shading).
(C) Sanger-sequenced RNA (copy DNA) transcripts suggest that the c.634G>A allele is not detectable as a stable transcript, indicated by a single chromatogram peak at both the c.634G>A locus (red arrowhead) and the c.2176C>G variant (blue arrowhead) in the PR clone. The double chromatogram peak at the synonymous 3 bp substitution (gray) and the c. 2176C>G variant locus (not gene-corrected; blue) in the GC clone indicates that gene correction has rescued this transcripts.
(D) qPCR identifies 50% relative mRNA expression in PR iPSCs compared with control cells (data represent the mean + 95% CI; p = 0.0001).
(E) Capillary western blot reveals 43% relative IFT140 expression in PR differentiated renal epithelial cells compared with GC cells (data represent the mean + 95% CI; p < 0.001).
(F) Bright-field images of kidney organoids on day 25 of culture (day 18 of aggregate culture). Scale bars, 1 mm.
(G) Immunofluorescence images of whole organoids demonstrate balanced expression of glomerular precursors (NPHS1, white), proximal tubule, (LTL, blue), distal tubule (CDH1, green), and collecting duct (CDH1, green; GATA3, red). Scale bars, 1 mm (low-magnification images) and 50 μm (high-magnification images).