Figure 4.
RNA-Seq of Epithelial Sub-fractions of Organoids Demonstrates Dysfunctional Cellular Processes Resulting from IFT140 Variants
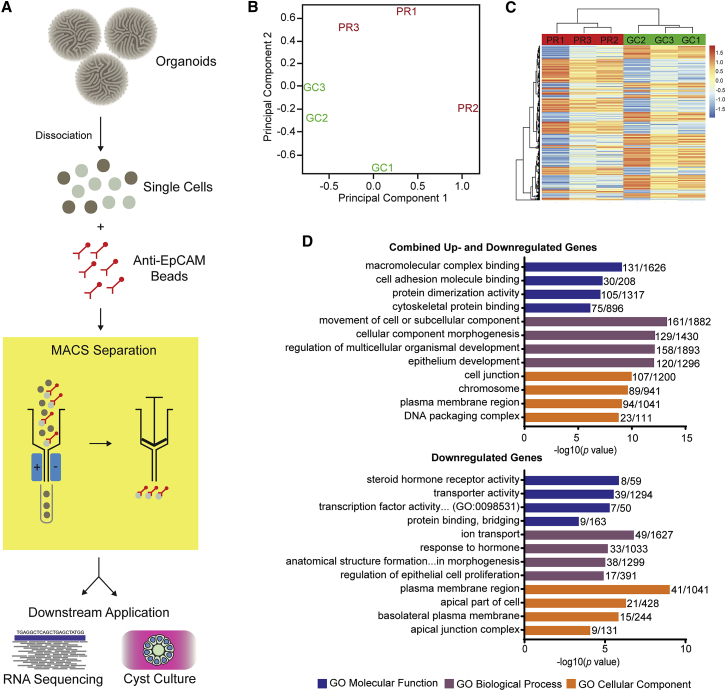
(A) Diagrammatic representation of epithelial MACS from dissociated organoids. Dissociated organoids were incubated with magnetic beads conjugated with EPCAM antibody and passed through a magnetic field, trapping epithelial cells that could be gently eluted for downstream application after removal of the magnetic field.
(B) Principal-component analysis of primary RNA-seq data demonstrates clustering of PR and GC replicates.
(C) Heatmap demonstrates consistency of gene expression across replicates.
(D) Top GO terms output from ToppGene gene-list enrichment of set 1 (combined up- and downregulated genes with significant differential expression, adjusted p < 0.01; top graph) and set 2 (genes with log fold change > 0.7 favoring GC and significant differential expression < 0.05; bottom graph). Bars represent the −log10(p value), and the adjacent fraction represents the number of genes within the dataset (numerator) and the number of genes within the GO gene list (denominator). Similar GO terms with a near-identical DGE representation and GO term were filtered out to avoid repetition. Colors are as follows: blue, GO molecular function; purple, GO biological process; and orange, GO cellular component.