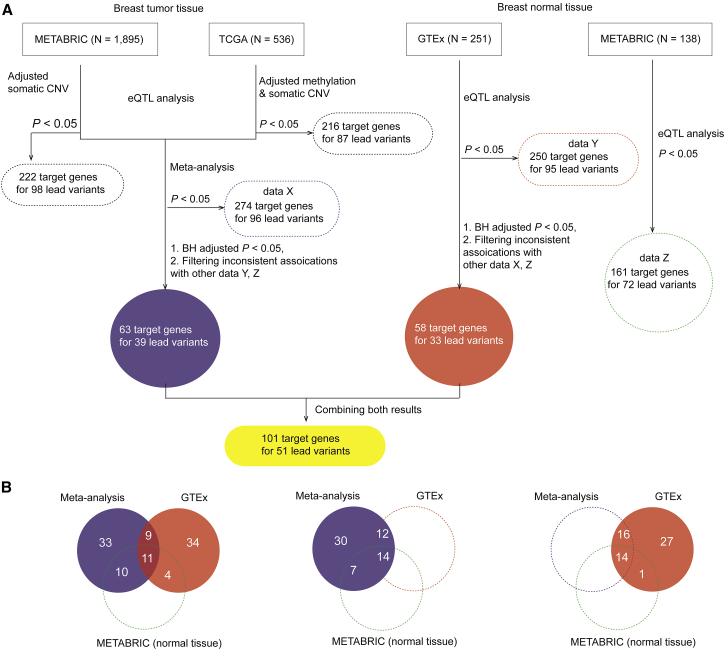
Figure 1.
A Workflow of Study Design
(A) A flow chart to illustrate the identification of target genes for GWAS lead variants based on cis-eQTL analysis using data from the METABRIC, TCGA, and GTEx datasets. The number in the dashed box indicates the total number of eQTL target genes that are identified by METABRIC, TCGA, GTEx, and meta-analysis at p < 0.05. The number in the highlighted blue and red box refers to the total of eQTL target genes that are identified using BH-adjusted p < 0.05 from a meta-analysis of tumor tissue results from METABRC and TCGA and normal tissues in GTEx, respectively. The number in the yellow box indicates the total number of genes after combing the results of the meta-analysis and the result in GTEx.
(B) The comparisons of the identified eQTL target genes with consistent associations across datasets (meta-analysis, GTEx, and normal tissues in METABRIC). From left to right, the number of the identified eQTL targets with consistent associations across the results from the meta-analysis and GTEx (BH-adjusted p < 0.05 for both) and normal tissue in METABRIC (unadjusted p < 0.05); from the meta-analysis (BH-adjusted p < 0.05), and GTEx and normal tissue in METABRIC (unadjusted p < 0.05 for both); from the meta-analysis (unadjusted p < 0.05), and GTEx (BH-adjusted p < 0.05) and normal tissue in METABRIC (unadjusted p < 0.05).