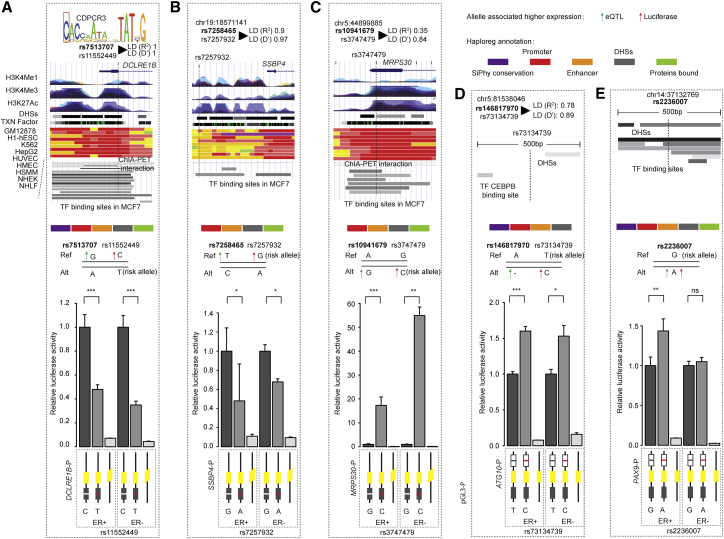
Figure 2.
Alternative Alleles Affecting Target Genes’ Promoter Activity
Alternative allele of functional SNPs rs11552449 (surrogate for rs7513707) (DCLRE1B), rs7257932 (surrogate for rs7258465) (SSBP4), rs3747479 (surrogate for rs10941679) (MRPS30), rs73134739 (surrogate for rs146817970) (ATG10), and rs2236007 (PAX9) affecting promoter activities of target genes. From left to right: rs11552449 (DCLRE1B) (A), rs7257932 (SSBP4) (B), rs3747479 (MRPS30) (C), - rs73134739 (ATG10) (D) and rs2236007 (PAX9) (E). At the top of each panel: the epigenetic landscape of functional SNPs. From top to bottom, functional SNP in LD mapped to TF motif (if exists); LD R2 value between lead SNP (bold) and functional SNP in European population; RefSeq genes; layered H3K4Me1, H3K4Me3, and H3K27Ac histone modifications; DNase clusters; clustered ChIP-seq binding sites; annotation using chromatin states on the ENCODE cell lines; ChIA-PET interactions in MCF-7 cell; and TF binding sites. The signals of different layered histone modifications from the same ENCODE cell line are shown in the same color (the detailed color scheme for each ENCODE cell line is described in the UCSC Genome Browser). The red in chromatin states refers to active promoter. For the ChIA-PET track, black lines represent interactions with the promoter region, and gray lines represent chromatin interactions that do not involve the promoter region. The corresponding location of the variant is indicated by a dashed line. The HaploReg annotation51 for each functional variant is indicated in the top right panel. At the center of each panel (gray shadow box): the allele associated with higher gene expression is indicated by the upward pointing green (eQTL analysis) or red (luciferase reporter assay) arrow. At the bottom of each panel: the alternative allele of functional SNPs changing promoter activities using luciferase reporter assays in the ER+ MCF-7 and ER− SK-BR3 breast cancer cell lines. The fragment containing the reference allele of each SNP was cloned downstream for luciferase construct and an alternative allele was engineered into it. The alternative and reference alleles are indicated by the red and white lines, respectively. The error bars represent the standard deviation of promoter activities of target genes. A paired t test was performed to derive p value for each candidate functional SNP.