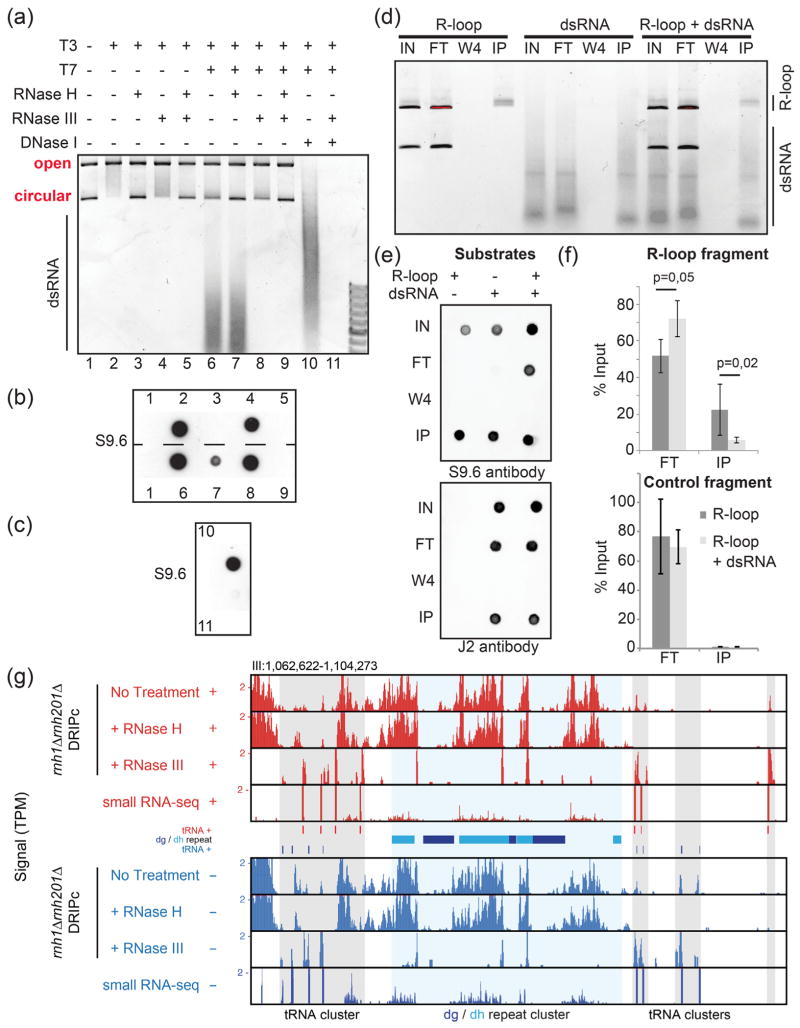
Fig. 2.
The S9.6 antibody recognizes and immune-precipitates dsRNAs both in vitro and in vivo in fission yeast. (a) A plasmid carrying the R-loop-forming AIRN mouse gene [17] was transcribed in vitro in the sense (T3) or antisense (T7) orientations as indicated. Soluble RNAs and proteins were then digested with RNase A and Proteinase K, respectively. After concomitant sense and antisense transcription, the reaction was split into two. In one half of the reaction, the plasmid was digested using DNase I treatment and only the dsRNAs were kept. (a) The reactions were run on an agarose gel. R-loops formed upon sense transcription (T3) induced a topological shift in the plasmid as reported previously [17]. When both T3 and T7 polymerases were used at the same time, RNase III-sensitive dsRNAs appeared in the gel. (b) Dot blot analysis of the above reactions using the S9.6 antibody. Upon sense and antisense transcription, the RNase H treatment is not sufficient to remove the signal. The signal fully disappears only after treatment with both RNase H and RNase III, suggesting that the S9.6 can recognize dsRNA in vitro. This was confirmed using the purified dsRNA (c). (d) R-loops and dsRNA were produced from the pFC53 plasmid. After transcription, the R-loop containing plasmids were cut with ApaLI to produce 2 bands, the longest of which contains R-loops. The smallest band acts as negative control because it does not carry R-loops. The cut plasmids and dsRNAs were then incubated alone or in combination with 4 μg of purified S9.6 antibody. The input (IN), corresponding to half of what was incubated with the antibody; the flow-through fraction (FT), corresponding to the material that did not bind to the antibody; the W4; and the immuno-precipitated fraction (IP) were then purified using phenol/chloroform extraction and ethanol precipitation and either run on an agarose gel (d) or analyzed by dot blot with the indicated antibodies (e). (f) The long fragment where R-loops can form (R-loop fragment) and the short fragment where R-loops do not form (control fragment) were also quantified by qPCR (n = 4 independent experiments, P-values with Wilcoxon–Mann–Whitney). The S9.6 antibody can immuno-precipitate GC-rich dsRNAs, and when dsRNAs are mixed with R-loops, the recovery of R-loops by the S9.6 antibody is reduced (see text). (g) Representative centromeric region of chromosome III showing RNase H-resistant but RNase III-sensitive DRIPc-seq signals at the dg and dh centromeric repeats. Note that most DRIPc-seq signals over tRNA genes are RNase H-sensitive, confirming that genuine R-loops form at tRNA genes [18].