Kadelka et al. show that parameters linked with HIV-1 broadly neutralizing antibody (bnAb) development shape HIV-1–binding antibody responses in an antigen and IgG subclass dependent manner. Identified HIV-1 antibody signature landscapes reveal a shift toward IgG1-driven responses in bnAb developers.
Abstract
Understanding pathways that promote HIV-1 broadly neutralizing antibody (bnAb) induction is crucial to advance bnAb-based vaccines. We recently demarcated host, viral, and disease parameters associated with bnAb development in a large HIV-1 cohort screen. By establishing comprehensive antibody signatures based on IgG1, IgG2, and IgG3 activity to 13 HIV-1 antigens in 4,281 individuals in the same cohort, we now show that the same four parameters that are significantly linked with neutralization breadth, namely viral load, infection length, viral diversity, and ethnicity, also strongly influence HIV-1–binding antibody responses. However, the effects proved selective, shaping binding antibody responses in an antigen and IgG subclass–dependent manner. IgG response landscapes in bnAb inducers indicated a differentially regulated, IgG1-driven HIV-1 antigen response, and IgG1 binding of the BG505 SOSIP trimer proved the best predictor of HIV-1 neutralization breadth in plasma. Our findings emphasize the need to unravel immune modulators that underlie the differentially regulated IgG response in bnAb inducers to guide vaccine development.
Introduction
Broadly neutralizing antibodies (bnAbs) to HIV-1 are defined by their superior activity against genetically diverse HIV-1 strains, referred to as neutralization breadth. Elite bnAbs that pair breadth with extraordinary potency are currently developed as lead components of vaccines and therapeutics (Burton and Hangartner, 2016; Caskey et al., 2016; Escolano et al., 2017; Pegu et al., 2017). bnAb responses are rare (10–25%) in natural infection, with highly potent elite bnAbs only detected in an estimated 1% of HIV-1 infections (Burton and Hangartner, 2016; Moore and Williamson, 2016; Rusert et al., 2016), and no vaccine thus far has succeeded in inducing bnAb activity (Burton and Hangartner, 2016; Escolano et al., 2017). Key elements that restrict and promote bnAb induction in natural HIV-1 infection thus need to be unraveled and exploited to overcome barriers in vaccine development. This includes the search for distinct HIV-1 Envelope (Env) variants that prime and mature bnAb responses (Wu et al., 2010; Sanders et al., 2015; Jardine et al., 2016; Stamatatos et al., 2017; Ward and Wilson, 2017) and a quest to define host, viral, and disease factors that stimulate bnAb development (Haynes et al., 2016; Landais et al., 2016; Moore and Williamson, 2016; Rusert et al., 2016; Borrow and Moody, 2017). Based on cohort studies and in-depth analyses of bnAb and virus coevolution in individual patients, various parameters have been implicated as drivers of bnAb induction. These include exposure to high loads of viral antigen over several years of infection and a high viral diversity as a result of virus evolution or superinfection (Sather et al., 2009; Cortez et al., 2012; Doria-Rose et al., 2014; Goo et al., 2014; Hraber et al., 2014; Bhiman et al., 2015; Landais et al., 2016; Rusert et al., 2016). In addition, a range of immune parameters including peripheral CD4 cell levels, distinct regulatory CD4 T cell subsets (Moody et al., 2016; Borrow and Moody, 2017), soluble CXCL13 (Havenar-Daughton et al., 2016; Dugast et al., 2017), and specific HLA variants (Landais et al., 2016) have been implicated in bnAb evolution.
To decipher factors that promote bnAb activity and their individual influence we recently conducted a systematic survey of bnAb activity in 4,484 chronically HIV-1–infected individuals (Rusert et al., 2016). This population-wide screen, which we now refer to as the Swiss 4.5K Screen, identified 239 patients as bnAb inducers and discerned four main parameters significantly associated with bnAb development: infection length, viral load, viral diversity, and black ethnicity. Here, building on the Swiss 4.5K Screen, we generate a landscape of HIV-1 Env and Gag binding antibody responses in the same patient cohort to define immune regulatory principles that underlie the link between the identified factors and bnAb evolution. In particular, we seek to (1) delineate parameters that steer HIV-binding antibody responses, (2) define if and how these parameters intersect with factors that steer bnAb development, (3) investigate shifts in the HIV antibody response landscape linked with the development of neutralization breadth, and (4) identify HIV-binding antibody signatures that define neutralization breadth.
Results
Systematic survey of factors that steer IgG subclass responses to HIV-1 Env and Gag antigens
A key aspect of bnAb evolution that awaits resolution is whether bnAbs are unique in the dependence on cofactors or whether these factors also drive other HIV-1–specific antibody responses. Building on the Swiss 4.5K Screen (Rusert et al., 2016), we performed a comprehensive survey of plasma IgG activity in 4,281 asymptomatic, viremic, chronic HIV-1–infected individuals (Fig. 1, A–G) to 13 HIV-1 antigens (Fig. 1 H), including Gag antigens p17 and p24 and several Env antigens comprising the closed, native-like BG505 trimer, open Env conformations (gp120 [JR-FL] and gp140 [BG505]), and distinct epitope regions (V3 loop, CD4-binding site [CD4bs], membrane proximal external region [MPER], and gp41ΔMPER; Fig. 1 I and Tables S1 and S2). The obtained serology data (Fig. 1 I) were then used to probe for interrelations of HIV-binding antibody activity with viral, host, and disease determinants and neutralization breadth (Fig. 1 A).
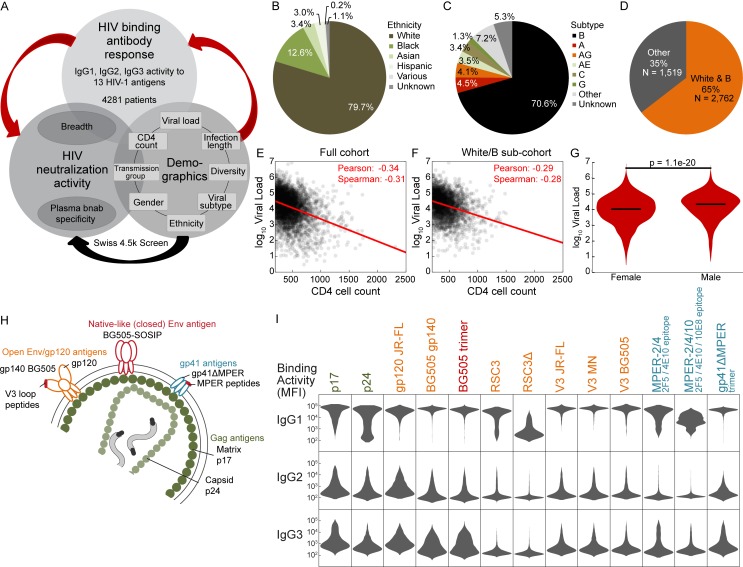
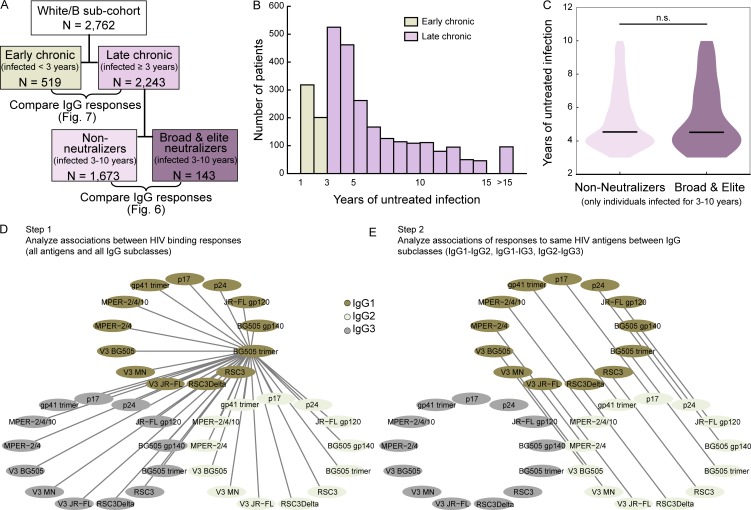
Figure 1.
Systematic survey of IgG subclass responses to HIV-1 Env and Gag antigens. (A) Experimental study design. Red arrows indicate aspects examined in the current study. (B and C) Distribution of ethnicities (B) and HIV-1 subtypes (C) in the cohort. (D) Relative size of the white/B subcohort. (E and F) Scatterplot of log10 viral load and CD4 cell count for the full cohort (E) and the white/B subcohort (F). A red linear regression line and the Spearman and Pearson correlation coefficients are shown. (G) Distribution of log10 viral load in females and males in the full cohort (pMann-Whitney = 1.1e-20). (H) Overview of 13 HIV-1 antigens selected for IgG response monitoring. (I) Summary of measured IgG-binding antibody response (MFI raw data; Luminex binding assay; single measurements) in 4,281 chronic HIV-1–infected individuals. Relative binding activities (Table S1) derived from these MFI raw data were used for further analysis.
HIV-1 IgG subclass responses can differ dependent on the HIV antigen type, disease progression, and immunization regimen (Klasse et al., 1990; Lal et al., 1991; Thomas et al., 1996; Binley et al., 1997; Ngo-Giang-Huong et al., 2001; Voltersvik et al., 2003; Martinez et al., 2005; Banerjee et al., 2010; French et al., 2013; Lai et al., 2014; Yates et al., 2014; Chung et al., 2015; Ackerman et al., 2017). Here, we established a landscape of HIV-1 IgG subclass responses in chronic HIV-1 infection across Gag and Env antigens, focusing on IgG1, IgG2, and IgG3, as HIV-1 IgG4 responses are known to be low (Broliden et al., 1989; Banerjee et al., 2010). In line with the known dominance of IgG1 in HIV-1 infection (Broliden et al., 1989; Binley et al., 1997; Banerjee et al., 2010), IgG1 responses showed the highest reactivity across all antigens, followed by IgG3 and IgG2 responses (Fig. 1 I). Using the collected binding data, we used multivariable linear regression models to assess how individual factors influence HIV-1 IgG responses. Results from univariable analysis are additionally provided to highlight which effects are most affected by other variables. We assessed eight parameters, for which we had complete datasets for 3,159 individuals: two patient characteristics (gender and ethnicity) and six viral and disease parameters (viral load and CD4 level at the time point of sampling, pol diversity, infection length, transmission mode, and HIV-1 pol subtype; Figs. 2 and 3; and Tables S2 and S3).
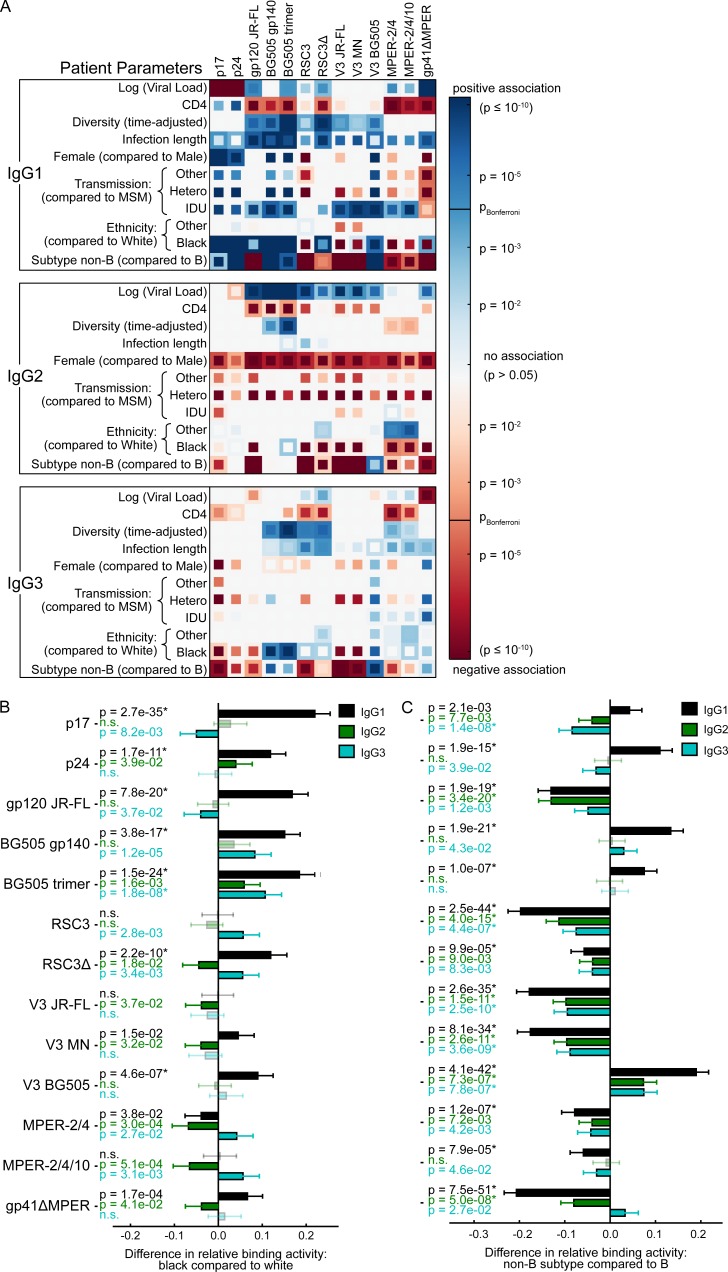
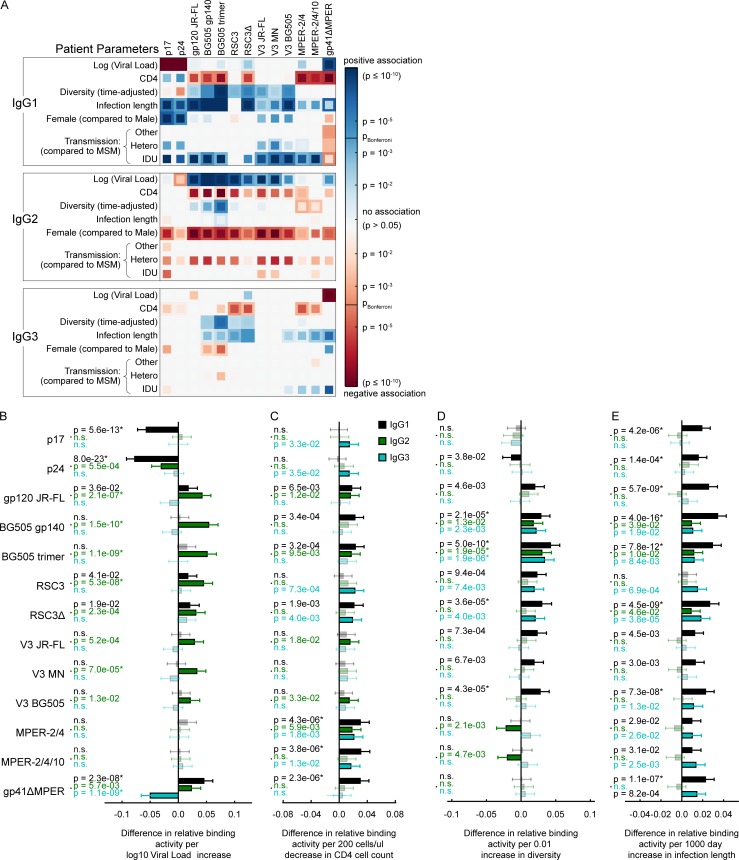
Figure 2.
Influence of host, viral and disease parameters on binding antibody responses across all ethnicities. (A) The influence of the virus load, CD4 count, viral diversity, length of untreated HIV-1 infection, transmission route, gender, ethnicity, and HIV-1 subtype on binding antibody responses to each tested HIV-1 antigen was determined by univariable (inner squares) and multivariable (outer squares) linear regression for all three IgG subclasses (n = 3,159). Only significant associations (P < 0.05) are colored, and the Bonferroni-corrected significance threshold (P = 0.00012) is shown in the color map. Color intensity represents the significance level of positive (blue) and negative (red) associations. See Table S3 for detailed regression results. (B and C) Impact of the indicated parameter on the relative IgG1 (black), IgG2 (green), and IgG3 (blue) binding activity, based on the same multivariable linear regression analysis as in A. Error bars depict the 95% confidence intervals. Nonsignificant associations are marked by n.s. and are shown in lighter color shades. * highlights significant associations when using a Bonferroni correction for multiple testing. (B) Influence of black compared with white individuals. (C) Non–B-infected compared with B-infected individuals.
Figure 3.
Influence of host, viral and disease parameters on binding antibody responses in the white/B subcohort. (A) The influence of the virus load, CD4 count, viral diversity, length of untreated HIV-1 infection, gender, and transmission route on binding antibody responses was determined by univariable (inner squares) and multivariable (outer squares) linear regression for each IgG subclass and each HIV-1 antigen (n = 2,122). Only significant associations (P < 0.05) are colored, and the Bonferroni-corrected significance threshold (P = 0.00016) is shown in the color map. Color intensity represents the significance level of positive (blue) and negative (red) associations. See Table S3 for detailed regression results. (B–E) Impact of the indicated parameter on the relative IgG1 (black), IgG2 (green), and IgG3 (blue) binding activity, based on the same multivariable linear regression analysis as in A. Error bars depict the 95% confidence intervals. Nonsignificant associations are marked by n.s. and are shown in lighter color shades. * highlights significant associations when using a Bonferroni correction for multiple testing. (B) Impact of viral load per unit increase in log10 viral copy numbers per milliliter of blood. (C) Impact of CD4 cell count per 200 cells/µl decrease in CD4 cell count. (D) Impact of viral diversity per 0.01 increase in pol diversity. (E) Impact of infection length per 1,000 d increase in length of untreated HIV-1 infection.
Black ethnicity is linked with higher IgG1 responses to HIV-1 Env
A main driving factor of bnAb elicitation identified in the Swiss 4.5K Screen was black ethnicity (Rusert et al., 2016). Here we show that black ethnicity is significantly linked with elevated IgG1 Gag and several IgG1 Env responses, but not with IgG2 Env and only marginally with IgG3 Env (Fig. 2, A and B), suggesting the manifestation of an immune stimulatory environment that specifically promotes IgG1 responses.
White individuals in our cohort are mostly infected with HIV-1 subtype B, whereas non-B subtypes are predominantly represented within other ethnicities (Schoeni-Affolter et al., 2010; Rieder et al., 2011; Rusert et al., 2016; Table S2). Multivariable analysis revealed that the effect of ethnicity on HIV-binding antibodies was independent of the infecting subtype (Fig. 2, A and B). We did not attempt a direct assessment of the influence of the infecting subtype on binding antibody responses, as it is expected that in the binding assay, antibodies preferentially recognize antigens that are derived from the same subtype and that recognition of antigens from other subtypes can vary (Fig. 2, A and C). To formally exclude an influence of the analyte subtype in subsequent analyses, we examined, in addition to the full dataset, also the subcohort of white individuals infected with subtype B (white/B subcohort; n = 2,762) as indicated throughout our study (Fig. 3 and Table S2).
Differential influence of viral load, diversity, and infection length
Untreated HIV-1 infection is characterized by an inverse relationship of CD4 levels and viral loads (Mellors et al., 1997). Although this was also apparent in our cohort (Fig. 1, E and F), multivariable analysis paired with the large sample size enabled a dissection of individual influences of CD4 cell and viral load levels. Viral load portrayed a dichotomy by showing a highly significant, negative effect on IgG1 p17, IgG1 p24, and IgG3 gp41ΔMPER responses, contrasted by a positive association with IgG1 gp41ΔMPER and IgG2 gp120 responses, in particular the IgG2 BG505 trimer and BG505 gp140 reactivity (Fig. 2 A and Fig. 3, A and B). A differential regulation of Gag and Env antibody responses has been noted for long. In particular, low anti-Gag IgG responses have been suggested to be linked with disease progression, possibly reflecting the waning CD4 cell counts at later disease stages and potentially unveiling a higher dependence of Gag responses on CD4 help (Broliden et al., 1989; Klasse et al., 1990; Lal et al., 1991; Thomas et al., 1996; Binley et al., 1997; Ngo-Giang-Huong et al., 2001; Voltersvik et al., 2003; Trkola et al., 2004; Martinez et al., 2005; Tomaras and Haynes, 2009; Banerjee et al., 2010; French et al., 2010, 2013; Yates et al., 2014; Chung et al., 2015; Ackerman et al., 2016). Results from multivariable testing in our cohort, however, suggested otherwise, revealing that CD4 levels by themselves have a marginal effect on shaping HIV-binding antibody responses in asymptomatic chronic infection (Fig. 2 A and Fig. 3, A and C). Two observations were particularly noteworthy. First, IgG1 p17, IgG1 p24, and IgG3 gp41ΔMPER responses all exhibited a strong negative dependency on viral load, but were not significantly affected by CD4 levels when controlled for influence of other factors in the multivariable analysis. Second, the IgG1 responses to MPER and gp41ΔMPER were positively linked with lower CD4 counts.
Viral diversity, defined per pol sequence ambiguity, is strongly positively associated with bnAb development in our cohort (Rusert et al., 2016). Intriguingly, diversity also positively influenced gp120 responses (irrespective of the antigen subtype or its monomeric or trimeric state), but not Gag, MPER, or gp41ΔMPER responses (Fig. 2 A and Fig. 3, A and D). The effect was largely restricted to IgG1, but also notable for some IgG2 and IgG3 Env responses. Thus, in line with the higher genetic variability of gp120, antibody responses to gp120 appear to improve upon exposure to a higher viral diversity, whereas the responses to the genetically more conserved Gag, MPER, and gp41ΔMPER antigens remain largely unaffected. Remarkably, diversity had the strongest effect on BG505 trimer binding both in the full cohort and the white/B subcohort.
Infection length, which is also strongly positively linked with bnAb development in our cohort (Rusert et al., 2016), predominantly influenced IgG1 responses across Gag and Env antigens, whereas no or low influence was observed for IgG2 and IgG3 responses, respectively (Fig. 2 A and Fig. 3, A and E).
Patient demographic influences on the HIV-1 IgG response
We observed interesting patient demographic effects among females and intravenous drug users (IDUs). We previously noted that females developed neutralization breadth slightly less frequently than males in our cohort (Table S7 in Rusert et al., 2016). Here, we show that females exhibited higher p17 and p24 IgG1 responses (Fig. 2 A; Fig. 3 A; and Table S3). Both these responses were strongly negatively affected by viral load (Fig. 2 A and Fig. 3, A and B). However, although females are known to have lower viral loads than males (Sterling et al., 2001; Hagen and Altfeld, 2016), which was also the case in our study (Fig. 1 G), multivariable testing confirmed that the higher IgG1 p17 and p24 responses in females occurred independently of the viral load. Quite remarkable was the difference in IgG2 responses. Females developed less IgG2-binding antibodies across all 13 tested HIV-1 antigens (Fig. 2 A and Fig. 3 A), strongly suggesting specific influences on IgG2 class-switch pathways.
IDUs mounted significantly stronger IgG1 binding to various HIV-1 antigens in line with a previously suggested, differential IgG activity of IDUs (Bongertz et al., 1999), but showed no difference in IgG2 and IgG3 responses (Fig. 2 A and Fig. 3 A). Although triggers of the enhanced IgG1 response are likely different in IDUs, the observed pattern is largely reminiscent of what we observed in black individuals. Intriguingly, IDUs in our cohort develop neutralization breadth slightly more frequently (Table S7 in Rusert et al., 2016), which, in conjunction with the observations made for black individuals, suggests the existence of immune regulatory mechanisms that promote a dominant IgG1 pathway linked with neutralization breadth.
IgG1-driven HIV antibody response landscape is linked with the development of neutralization breadth
The predominance of IgG1-binding antibodies in individuals with black ethnicity and IDUs prompted us to investigate the influence of cofactors on the pattern of IgG subclass responses. Comparing the relative binding activities between IgG subclasses (IgG1-IgG2, IgG1-IgG3, and IgG2-IgG3) in the full cohort (Fig. 4, A–C) and the white/subtype B subcohort (Fig. 4, D–F) highlighted a predominant IgG1 response to several Env antigens that is linked with neutralization breadth (Fig. 4, G–I). Of note, high IgG1 activity to the BG505 trimer was a strong indicator of neutralization breadth as was a high IgG3 MPER activity.
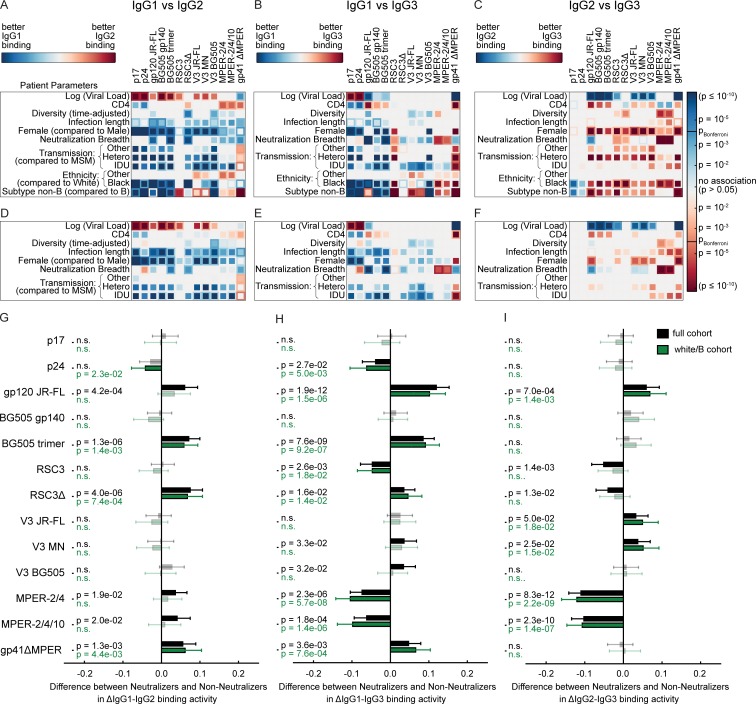
Figure 4.
Comparison of relative binding activities between IgG subclasses. (A–I) Analysis of the full cohort (n = 3,159; A–C and G–I) and the white/B subcohort (n = 2,122; D–I) with complete data on the included host, viral, and disease parameters. The influence of the virus load, CD4 count, viral diversity, length of untreated HIV-1 infection, transmission route, gender, neutralization breadth (at least cross neutralization), ethnicity, and HIV-1 subtype on the differences in binding antibody responses to each tested HIV-1 antigen was determined by univariable (inner squares) and multivariable (outer squares) linear regression for all three pairs of IgG subclasses. Only significant associations (P < 0.05) are colored, and the Bonferroni-corrected significance threshold (P = 0.00012) is shown in the color map. The color intensity represents the significance level. (G–I) Difference in neutralizers and nonneutralizers in differential IgG subclass (ΔIgG) binding activity, based on the same multivariable linear regression analysis as in A–F. Error bars depict the 95% confidence intervals. Nonsignificant associations are marked by n.s. and are shown in lighter color shades.
To investigate global shifts in antibody response landscapes that are linked with neutralization breadth, we compared correlation networks of binding responses of nonneutralizers and potent neutralizers (individuals with previously defined broad and elite neutralization activity [Rusert et al., 2016]). To enable an unbiased evaluation, we restricted the analysis to the white/B subgroup and therein to individuals with chronic HIV-1 infection (3–10 yr untreated infection) to ensure comparable infection length in neutralizers and nonneutralizers as detailed in the methods section. The accordingly defined subcohorts (Fig. 5, A–C; and Table S2) of nonneutralizers (n = 1,341; Fig. 6 A) and potent neutralizers (broad and elite activity; n = 95; Fig. 6 B) were then subjected to a correlation network analysis (Fig. 5 D). Potent neutralizers showed significant shifts in correlations for the majority of antigens both within the same IgG subclass and across subclasses (Fig. 6 C). Focusing on in-between subclass associations of responses to each antigen (Fig. 5 E), we detected significantly lower associations between IgG1/IgG2 and IgG1/IgG3 responses, but not between IgG2/IgG3 in potent neutralizers (Fig. 6 D), which was driven by an increase in IgG1 responses (Fig. 4). Of note, MPER responses constituted the only exception to the IgG1 driven shift in neutralizers.
Figure 5.
Distribution of untreated infection length and set-up of correlation network analyses. (A) Patient selection for the antibody response landscape analyses in Figs. 6 and 7. (B) Distribution of untreated infection length in the white/B subcohort. Colors distinguish early (n = 519) and late (n = 2,243) chronic infection and indicate subcohorts of early and late infection analyzed in Fig. 7. All samples with an untreated infection of >15 yr are grouped together (n = 96). (C) Distribution of untreated infection length in the white/B subcohort in individuals with 3- to 10-yr untreated infection and with no or weak neutralization activity (left; n = 1,341) or potent (broad or elite) neutralization activity (right; n = 95). The two distributions do not differ significantly (pMann-Whitney = 0.70). (D) Example of the correlation network analyses shown in Figs. 6 (A–C) and 7 (A–C). The relative binding activities to each antigen and each IgG subclass were compared with all other relative binding activities, as exemplified for IgG1 BG505 trimer binding. (E) Example of the correlation network analysis as conducted in Figs. 6 D and 7 D. Only the correlations between the IgG1 and IgG2 responses of the same antigen were investigated. Correlations for IgG1/IgG3 and IgG2/IgG3 responses were conducted the same way.
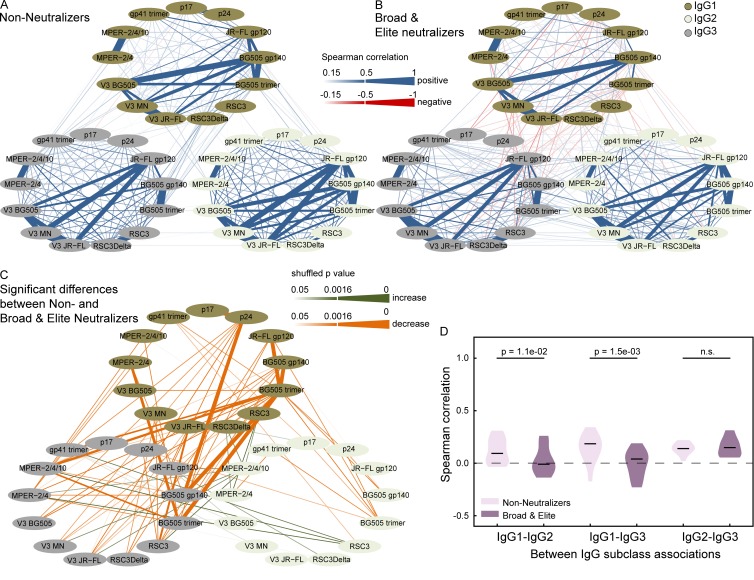
Figure 6.
Differential landscape of HIV-1 IgG responses in broad neutralizers. (A and B) Pairwise Spearman correlation analysis of the relative binding activities to the 13 HIV-1 antigens within and between subclass IgG1 (dark green), IgG2 (light green), and IgG3 (gray) in white subtype B–infected patients with late chronic (3- to 10-yr untreated) infection. Correlations with a magnitude >0.15 are depicted in blue (positive associations) and red (negative associations). The strength of Spearman correlation is signified by color intensity for correlations with magnitudes from 0.15 to 0.5 and additionally by line width for magnitudes >0.5. (A) Patients with no or low neutralizing activity (n = 1,673). (B) Patients with broad or elite neutralizing activity (n = 143). (C) Significant differences (pshuffle < 0.05; 10,000 random reshuffles) in pairwise Spearman correlations between broad neutralizers (as in B) and nonneutralizers (as in A) are shown. Green and orange shaded lines denote correlations that increase or decrease among broad neutralizers, respectively. The significance level of differences is depicted by color intensity and additionally by line width for differences that remain significant after correction for multiple testing (Benjamini-Hochberg correction with a false discovery rate of 10%; pshuffle ≤ 0.0016). (D) The distribution of all pairwise between IgG subclass correlations of individual antigens (Fig. 5 E) is shown for the nonneutralizers (light violet) and potent (broad or elite) neutralizers (dark violet). Significant p-values from a paired Wilcoxon test are shown.
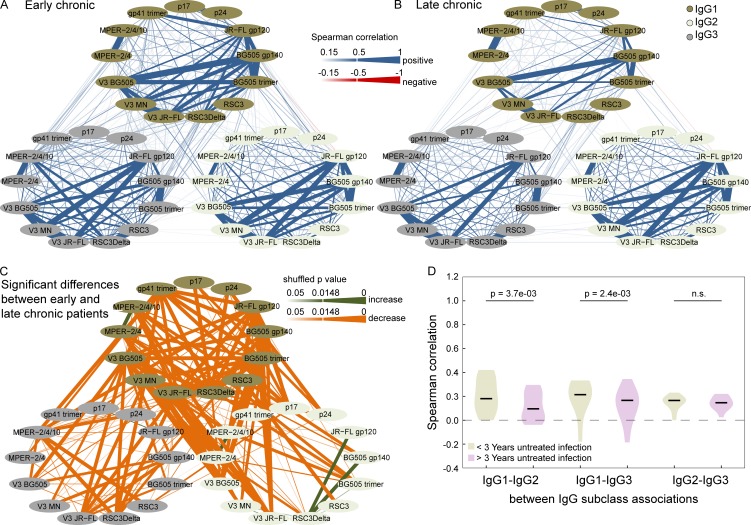
Importantly, we show that this IgG1-driven antibody response landscape develops with disease progression increasing from early to late HIV infection (Fig. 7), lending further support that the widely observed late development of bnAbs (after three or more years of untreated infection) is tied to immune regulatory changes.
Figure 7.
Correlations of antibody binding responses in early and late chronic infection. (A and B) Pairwise Spearman correlation analysis of the relative binding activities to the 13 HIV-1 antigens within and between subclass IgG1 (dark green), IgG2 (light green), and IgG3 (gray) in white subtype B infected patients with early chronic (1–3 yr untreated infection; n = 519; A) and late chronic (>3 yr untreated infection; n = 2,243; B) infection. Correlations with a magnitude >0.15 are depicted in blue (positive associations) and red (negative associations). The strength of Spearman correlation is signified by color intensity for correlations with magnitudes from 0.15 to 0.5 and by the strongest color intensity and line width for magnitudes >0.5. (C) Significant differences (pshuffle < 0.05; 10,000 random reshuffles) in pairwise Spearman correlations between early chronic (as in A) and late chronic (as in B) patients are shown. Green and orange shaded lines denote correlations that increase or decrease among late chronic patients, respectively. The significance level of differences is depicted by color intensity and by line width for differences that remain significant after correction for multiple testing (Benjamini-Hochberg correction with a false discovery rate of 10%; pshuffle ≤ 0.0148). (D) The distribution of all pairwise “between IgG subclass” correlations of individual antigens (Fig. 5 E) is shown for the early chronic (green) and the late chronic (violet) patients. Significant p-values from a paired Wilcoxon test are shown.
Besides the observed global shifts in IgG subclass associations between nonneutralizers and neutralizers (Fig. 4), the correlations among IgG1 responses also markedly differed (Fig. 6). Nonneutralizers displayed a strong correlation between IgG1 activity to the BG505 trimer and several open Env antigens (BG505 gp140, V3 BG505, and JR-FL gp120) that was considerably lower in neutralizers, highlighting a qualitative and quantitative shift in the IgG1 anti-Env responses between the two groups.
Immune signatures linked with neutralization breadth
To distinguish HIV-binding antibody signatures that define neutralization breadth, we compared HIV IgG reactivity in patient groups with elite, broad, and cross neutralization activity to individuals with no or low neutralization activity (Rusert et al., 2016). To minimize potential confounders, we focused this analysis on the white/B subcohort. Importantly, all relevant findings remained valid in the full cohort (Table S4). A first visual comparison of the relative IgG reactivity for each antigen and IgG subclass indicated that high reactivity to certain antigens is more frequent among individuals with cross, broad, and elite neutralization activity (Fig. 8 A). Intriguingly, the most evident difference between bnAb developers and nonneutralizers was in the activity against the BG505 trimer consistent with the concept that trimer binding is a prerequisite for neutralization (Sattentau and Moore, 1995; Yasmeen et al., 2014; Sanders et al., 2015; Burton and Hangartner, 2016).
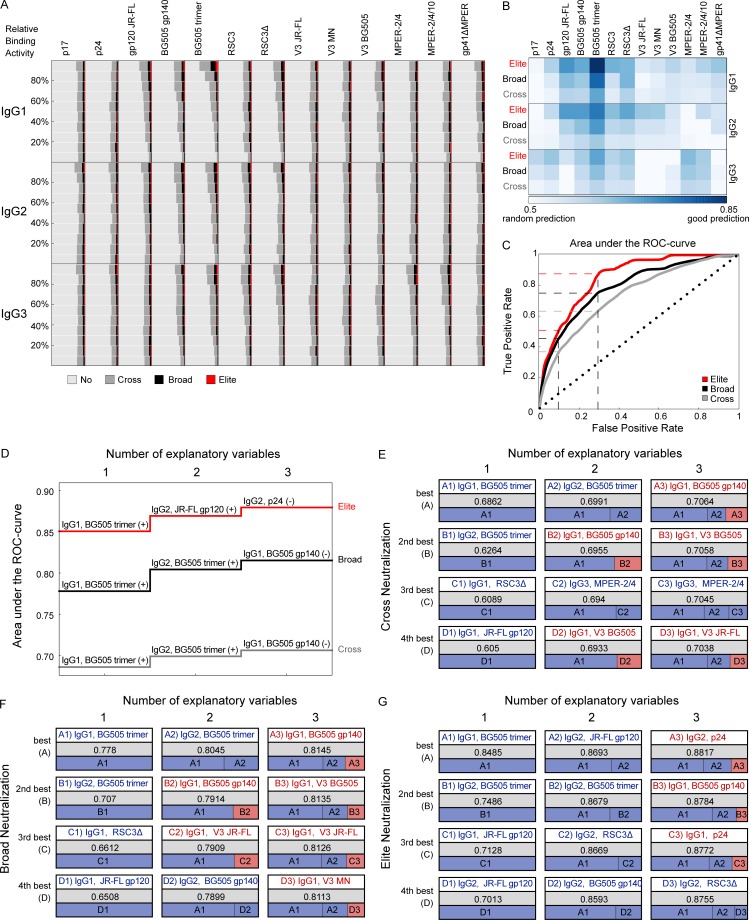
Figure 8.
HIV-1–binding antibody responses predict bnAb activity. Analysis of white/B subcohort (n = 2,762). (A) Frequency comparison of antigen reactivity in patient groups with no or weak- (n = 2,155; light gray), cross- (n = 453; dark gray), broad- (n = 123; black), and elite- (n = 31; red) neutralizing activity (Table S2). For each antigen and each IgG subclass, plasma samples were distributed into ten groups based on their relative binding activity, and the distribution of neutralizers and nonneutralizers across these groups is depicted. (B and C) Relative binding activities were used in logistic regression models to predict whether a plasma with known elite- (red), at least broad- (broad and elite activity: black) or at least cross- (cross, broad, and elite activity: dark gray) neutralizing activity was part of the respective group. The AUC of the ROC using fivefold cross validation and 100 repetitions was used to measure predictive strength. (B) Comparison of the predictive strength (shades of blue based on AUC values) of all single binding antibody responses. (C) ROC curve using IgG1 BG505 trimer binding as predictor. Dashed lines indicate the respective true positive rates of cross, broad, and elite neutralization at fixed false positive rates of 0.1 and 0.3. The black dotted line y = x indicates the expected performance of a random prediction. (D) Best multivariable prediction model for cross, broad, and elite neutralization. The best model with a certain number of variables (up to three) is learned in a greedy fashion and depicted. (+) indicates antibody-binding responses with a positive parameter in the model (high binding responses are predictive of neutralization breadth). (−) indicates negative parameters (low-binding responses are predictive of neutralization breadth). (E–G) At each step, the four best choices are shown from top (best) to bottom (fourth best). The respective AUC values and a representation of the impact of each binding activity on the respective model are also depicted. Antibody binding responses indicated in blue have a positive parameter in the model; red denotes a negative parameter. (E) Best prediction for cross neutralization. (F) Best prediction for broad neutralization. (G) Best prediction for elite neutralization. See Table S4 for AUC values and comparable findings for the full cohort.
Quantification of the predictive strength of antigen binding on HIV-1 neutralization by the area under curve (AUC) of the receiver-operating characteristic (ROC) disclosed a wide range of HIV IgG reactivity with predictive power toward identifying neutralization breadth (Fig. 8, B and C; and Table S4). IgG1 binding to the BG505 trimer proved to be the best single indicator of neutralizing activity with AUC values of up to 0.849, followed by IgG2 BG505 trimer and JR-FL gp120 (AUCs of up to 0.749 and 0.713, respectively). Solely based on IgG1 BG505 trimer binding activity 50.9% of elite, 44.0% of broad, and 27.6% of cross neutralizers in the white/B subcohort could be identified at a false positive rate of 10% (Fig. 8 C). Accepting 30% false positives, these numbers increased to 89.5%, 78.1%, and 57.8%, respectively. To define HIV immune signatures that maximize neutralization breadth predictions, we next established logistic-regression based models on multiple binding activities. Starting with the IgG1 BG505 trimer, the model was expanded in a greedy fashion by iteratively adding the predictor with the best predictive power (Fig. 8 D). Interestingly, the predictive power saturated quickly, and the four best choices for the second and third variable yielded similar predictive values (Fig. 8, E–G). Across elite, broad, and cross neutralization, a common three-variable immune signature for neutralization prediction emerged, characterized by high reactivity with a closed Env conformation (IgG1 and IgG2 BG505 trimer), but lower reactivity with an open Env conformation (IgG1 BG505 gp140), validating the recognized importance of trimer binding for neutralization activity (Sattentau and Moore, 1995; Yasmeen et al., 2014; Sanders et al., 2015; Burton and Hangartner, 2016). Notably, several IgG2 responses were among the best predictors in these multiparameter prediction signatures (Fig. 8, D–G), supporting a role of IgG2 responses as surrogate markers also in neutralization breadth development (Lal et al., 1991; Ngo-Giang-Huong et al., 2001; Martinez et al., 2005; French et al., 2010, 2013).
Immune signatures linked with neutralization specificity
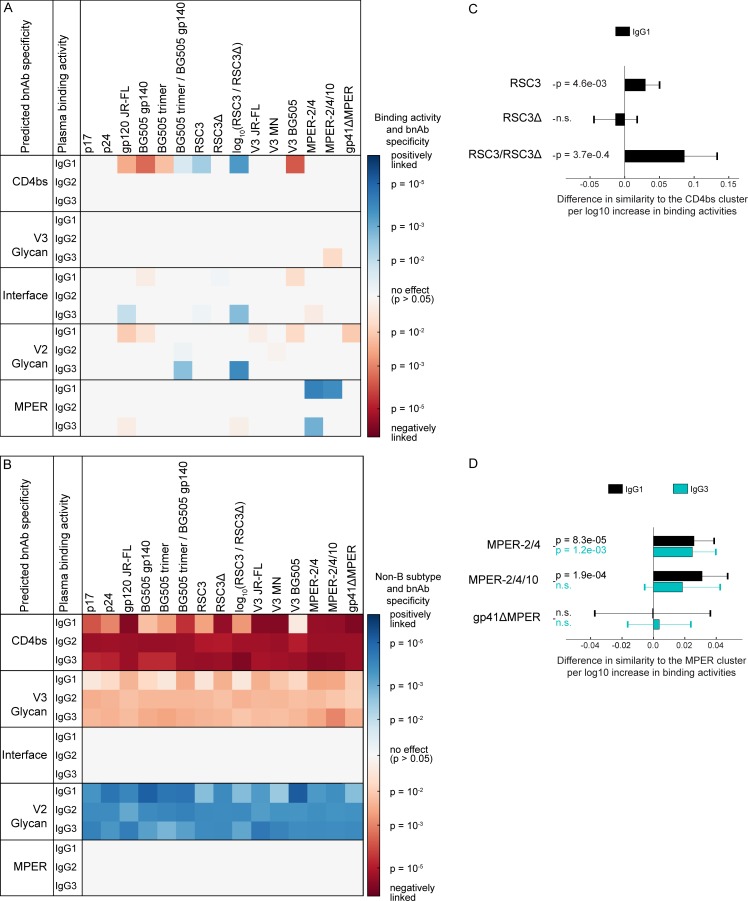
To probe for immune signatures that are linked with certain bnAb specificities, we investigated binding antibody patterns in 105 bnAb plasma samples, for which epitope specificity was previously predicted by neutralization fingerprint analysis (Fig. 9 and Table S5; Rusert et al., 2016). For two bnAb types, CD4bs and MPER, specific antigens (RSC3/RSC3Δ and MPER peptides, respectively) were included in our screen and thus allowed us to quantify the impact of epitope-specific binding in predicting bnAb types. Many antibodies targeting the CD4bs, including the bnAb VRC01, strongly react with RSC3 but not RSC3Δ, which lacks a crucial isoleucine at position 371 (Wu et al., 2010). Accordingly, the neutralization fingerprint similarity to CD4bs bnAbs in our cohort was linked with enhanced IgG1 binding to RSC3 and a higher ratio of IgG1 RSC3 to RSC3Δ binding (Fig. 9, A and C), confirming the validity of the neutralization fingerprint and highlighting the presence of VRC01-like bnAbs. Intriguingly, the neutralization fingerprint similarity to MPER bnAbs was significantly associated with both IgG1 and IgG3 MPER binding with IgG1 yielding an even stronger effect (Fig. 9, A and D).
Figure 9.
HIV-1–binding antibody responses associated with specific bnAb types. (A and B) Definition of binding patterns that predict bnAb types. The association of two variables, the log10 IgG binding activities (single measurements; A), and the infecting subtype (B vs. non-B) on various bnAb specificities (B) was determined by multivariable linear regression, using the Spearman neutralization fingerprint similarity with bnAb clusters of the top neutralizing plasma samples (n = 105) as defined in Rusert et al. (2016) as a marker of bnAb specificity. Significant associations (P < 0.05) are colored, with positive and negative associations indicated in blue and red, respectively. The color intensity represents the significance level. (C and D) Magnitude and significance of associations between the IgG response to CD4bs antigens and CD4bs bnAb activity (C) and MPER antigens and MPER bnAb specificity (D), based on the same multivariable linear regression analysis as in A and B. Error bars depict the 95% confidence intervals. Nonsignificant associations (P > 0.05) are marked by n.s.
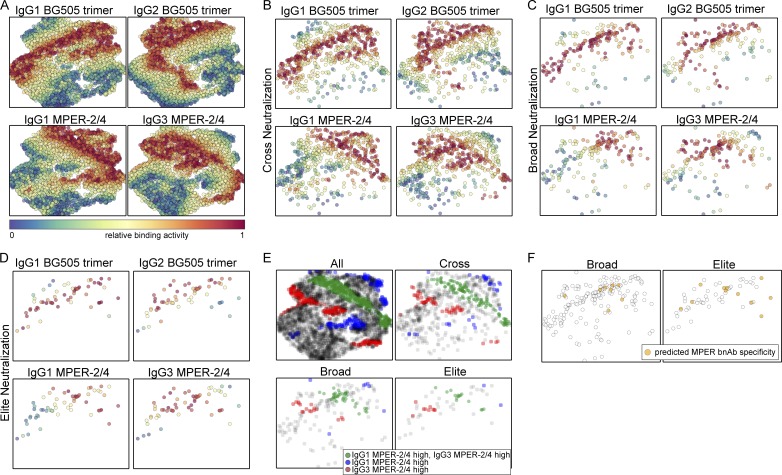
To further explore the association of IgG1 and IgG3 MPER binding responses with MPER targeting bnAb activity, we selected four markers, the two strongest positive markers of breadth (IgG1 and IgG2 BG505 trimer, which does not include the MPER domain) and the strongest MPER bnAb predictors (IgG1 and IgG3 MPER 2/4) and used this data to generate two-dimensional maps of the full cohort using the dimensionality reduction algorithm t-SNE (Fig. 10 A) and stratified according to the plasma neutralization activity (Fig. 10, B–D). Visualization of the IgG1 and IgG3 MPER single- and double-positive population (Fig. 10 E) side by side with plasmas with predicted MPER bnAb activity (Fig. 10 F; Rusert et al., 2016) in the same t-SNE plots revealed that 18 of 19 predicted MPER bnAbs fell into regions with MPER responses that were either dominant IgG3, dominant IgG1, or had a high IgG1 and IgG3 MPER reactivity, suggesting that MPER bnAb evolution should not be restricted to IgG3.
Figure 10.
Two-dimensional representation of all plasma samples based on binding antibody responses. Two-dimensional t-SNE map (Barnes-Hut-SNE approximation with 1,000 iterations) of all plasma samples (n = 4,281) based on the relative binding activities to BG505 trimer (IgG1 and IgG2) and MPER-2/4 (IgG1 and IgG3). (A) Points in each subplot are colored by the strength of response to one of the four relative binding activities (red color denotes high binding activity, and blue denotes low binding activity). (B–D) The t-SNE map from A is stratified by neutralization capacity, cross (n = 684; C), broad (n = 178; D), and elite (n = 58; E) neutralization. (E) The t-SNE map from A is stratified by neutralization capacity of plasma (all, cross, broad, or elite neutralization). Plasma with high relative binding (>0.667) for either IgG1 (blue) or IgG3 (red) MPER-2/4 or both (green) is colored. (F) The t-SNE map from A is stratified by neutralization capacity of plasma (broad or elite neutralization). Plasma with predicted MPER specificity (n = 19) is colored.
Discussion
The identification of immune regulatory pathways that are critical for bnAb elicitation by vaccines ultimately depends on decoding immune profiles linked with bnAb development in natural infection. Here, we applied a systems serology approach to retrieve HIV-1 antibody–binding signatures in 4,281 participants of the Swiss 4.5K Screen to define markers of bnAb induction. Exploring factors that steer HIV-1–binding antibody reactivity, we found that the same four parameters that are significantly linked with bnAb breadth in our cohort (Rusert et al., 2016), namely viral load, infection length, viral diversity, and ethnicity, also strongly influence HIV-1–binding antibody responses. Yet, their effect proved selective, shaping antibody responses in an antigen- and IgG subclass–dependent manner. This results in complex antibody immune signatures that are driven by combinations of parameters.
Based on the known antigenicity of the BG505 trimer, bnAbs are expected to preferentially bind this probe (Yasmeen et al., 2014). Interestingly, among the factors that prominently influence bnAb breadth, diversity, and infection length, but not viral load, were strongly positively linked with BG505 trimer IgG1 reactivity, suggesting that antigen quantity may not be the rate-limiting factor in the development of trimer-reactive antibodies. Infection length and viral diversity influenced the majority of IgG1 responses to gp120 antigens, but not MPER antigens. Likewise, there was no independent effect of infection length on IgG1 activity to the CD4bs specific probe RSC3. In sum, these results suggest that dependent on the desired antibody specificity vaccine strategies may need to simulate a different set of parameters.
This also includes the role of CD4 cells. Low CD4 levels have been suggested to foster potent bnAb development in some cohorts monitored for bnAb activity (Gray et al., 2011; Landais et al., 2016), but not in others (Piantadosi et al., 2009; Doria-Rose et al., 2010; Rusert et al., 2016). As CD4 levels and viral load are inversely correlated, and viral load strongly influences bnAb activity, assessing the independent contribution of CD4 levels to antibody evolution is critical. We previously showed that CD4 levels alone have no significant effect on the development of potent bnAb responses, but that, in agreement with earlier studies, less potent, cross-neutralizing activity is more frequently linked with lower CD4 levels (Rusert et al., 2016). Analyzing the same cohort of patients, we show now that CD4 levels have no strong impact on shaping gp120-binding responses. In contrast, the gp41 response, in particular MPER reactivity, was strongly inversely linked with CD4 levels (Figs. 2 and 3). MPER bnAbs are frequently autoreactive and it has been suggested that their evolution may benefit from a CD4-impaired environment with reduced tolerance control mechanisms (Haynes et al., 2005; Chen et al., 2013; Zhang et al., 2016; Borrow and Moody, 2017; Kelsoe and Haynes, 2017). The pronounced effect of low CD4 levels on gp41/MPER binding antibodies but not on potent bnAb development in our cohort suggests that employment of tolerance-modulating interventions to boost bnAb activity needs to be carefully balanced. Based on our results, the majority of bnAb types are unlikely to benefit substantially from a CD4 low environment but this could remain a possibility in the context of MPER vaccination regimens.
Females in our cohort developed slightly less neutralization breadth (Rusert et al., 2016). We discovered intriguing differences in the IgG reactivity pattern of females with elevated IgG1 p17 and p24 responses and a generally decreased IgG2 response. This may support a scenario of higher expression of X chromosome–linked regulators of the immune response (Fish, 2008; Hagen and Altfeld, 2016). The diminished IgG2 binding antibodies across all 13 tested HIV-1 antigens suggest specific influences on IgG2 class-switch pathways. Of note, IFN-γ has been implicated in the development of IgG2 responses (Finkelman et al., 1988; Kawano et al., 1994; Vazquez et al., 2015). Lower IFN-γ levels in HIV-1–infected females, as recently reported (Yong et al., 2016), could thus potentially contribute to the reduced IgG2 response in females.
A key finding of our study is that IgG subclass responses are differentially influenced, leading to distinctive antibody response landscapes among neutralizers and nonneutralizers. Particularly notable is the shift to IgG1 over IgG2 and IgG3 responses in neutralizers (Fig. 4). IgG1 is known to generally dominate in HIV infection (Broliden et al., 1989; Binley et al., 1997; Tomaras and Haynes, 2009; Banerjee et al., 2010). Antibody class switching to the four human IgG subclasses occurs through differential temporal pathways that are steered by various cytokines and epigenetic factors (Stavnezer, 1996; Zan and Casali, 2015; Horns et al., 2016). Elevated IgG1 levels may therefore result from conditions that favor subclass switching to IgG1 (Horns et al., 2016), increased frequencies of IgG1 specific cells, and/or elevate levels of IgG1 secretion (Knox et al., 2017). Of note, the IgG1 response to BG505 trimer correlated more strongly with the IgG1 response to open Env constructs that reveal the V3 loop (V3 peptides, JR-FL gp120, and BG505 gp140) in nonneutralizers than in neutralizers (Fig. 6, A–C). Reactivity with open Env constructs was generally high in our cohort (Fig. 1 I), but it was not a specific trait of nonneutralizers (Fig. 8, A and B), suggesting that a qualitative and quantitative shift to IgG1 BG505 trimer reactivity in broad and elite neutralizers appears to be a distinguishing factor, rather than a higher reactivity with open Env in nonneutralizers.
The fact that both black ethnicity and IDU status were significantly linked with elevated HIV-specific IgG1-binding activity and neutralization breadth is highly intriguing and suggests that IgG1 driven environments may be triggered by different factors. Mechanistically, it will be important to define in forthcoming studies whether the shift toward IgG1 responses is a result of higher frequencies of IgG1 B cells and/or elevated antibody secretion and to determine the underlying causes to aid incorporation of relevant immune modulatory strategies in vaccine regimens.
Although our results generally advocate for a stimulation of an IgG1 environment to foster gp120-directed binding and neutralizing responses, demands on the vaccine regimen may differ depending on the desired correlates of protection. For example, induction of nonneutralizing, antibody-dependent cell-mediate cytotoxicity–inducing anti-gp120 IgG3 responses, which were considered critical in the outcome of the RV144 trial (Kim et al., 2015), may depend on other parameters and require differential tailored vaccination regimen.
IgG3 responses have also been implicated in the induction of MPER bnAbs. Most MPER bnAbs isolated to date are IgG3 (Muster et al., 1993; Stiegler et al., 2001; Huang et al., 2012). In support of the importance of IgG3 for MPER antibody development, neutralizers exhibited higher IgG3 than IgG1 MPER-binding responses (Fig. 4). We nevertheless also observed strong links with IgG1 MPER responses lending support to an alternative scenario, where MPER bnAbs may originate in linked IgG3–IgG1 switch pathways, suggesting that IgG1 MPER bnAbs should likewise exist.
The comprehensive binding antibody analysis we conducted allowed us to explore two further principle questions: Which binding antibody responses are best suited to (1) predict bnAb activity (Fig. 8) and (2) define bnAb specificity (Fig. 9)? BG505 trimer reactivity proved the best predictor of neutralization breadth. Remarkably, additional parameters added to the prediction signature did only marginally improve the predictive capacity (Fig. 8, D–G). Of note, the MPER epitope is not included in the soluble BG505 trimer probe; yet, despite the prevalence of MPER bnAb inducers in our cohort (19 of 82 bnAb inducers with defined specificity (Rusert et al., 2016), MPER binding reactivity did not significantly add to the bnAb prediction signature. Our data thus strongly suggests that bnAb activity (irrespective of the type of bnAb elicited) develops in patients that mount a polyclonal trimer-reactive response and that thus reactivity with the native trimer, as in our study exemplified by the BG505-SOSIP trimer, demarcates a wide spectrum of bnAb activities. We do not expect this to be a unique feature of the BG505-SOSIP trimer, but rather a feature shared by all antigens that resemble the intact trimer. It will be interesting to explore in forthcoming studies the differential properties of intact trimer immunogens in demarcating neutralization activity to explore ex vivo their potential in eliciting desired responses upon vaccination. Although BG505 trimer reactivity potently indicated breadth in our study, it was not a distinctive predictor of bnAb epitope specificity, which was best predicted by epitope-specific binding probes such as RSC3 for CD4bs bnAb activity and MPER peptide reactivity for MPER bnAb responses.
In sum, the IgG1-driven landscape of HIV specific responses in potent neutralizers we identify here unveils the need of creating a specific immune regulatory environment for the development of neutralization breadth. Deciphering the basis of these regulatory shifts will be important to determine the triggers of this bnAb-favoring environment in forthcoming HIV pathogenicity studies and vaccine trials and to explore how this environment can be induced and maintained in healthy individuals to make bnAb-based vaccines possible.
Materials and Methods
Study populations and ethics information
The current study encompassed 4,281 plasma samples from viremic, HIV-1–infected individuals previously included in a population-wide screen for HIV-1 neutralization breadth, now termed the Swiss 4.5K Screen (Rusert et al., 2016). A detailed description of sample/patient selection and study design of the Swiss 4.5K Screen has been described previously (Rusert et al., 2016). The current study was restricted to 4,281 of the 4,484 included in the Swiss 4.5K Screen (Rusert et al., 2016) based on sample availability. All analyzed plasma samples were derived from specimen stored in the biobanks of the Swiss HIV Cohort Study (SHCS) and the Zurich Primary HIV Infection Study (ZPHI). The SHCS is a prospective, nationwide, longitudinal, noninterventional, observational, clinic-based cohort with semiannual visits and blood collections, enrolling all HIV-infected adults living in Switzerland (Schoeni-Affolter et al., 2010). The SHCS, founded in 1988, is highly representative of the HIV epidemiology in Switzerland as it includes an estimated 53% of all HIV cases diagnosed in Switzerland since the onset of the epidemic, 72% of all patients receiving antiretroviral therapy in Switzerland, and 69% of the nationwide registered AIDS cases (Schoeni-Affolter et al., 2010; Yang et al., 2015). The SHCS is registered under the Swiss National Science longitudinal platform. Detailed information on the study is openly available on http://www.shcs.ch.
The ZPHI is an ongoing, observational, nonrandomized, single center cohort founded in 2002 that specifically enrolls patients with documented acute or recent primary HIV-1 infection (http://www.clinicaltrials.gov; ID NCT00537966; Rieder et al., 2011).
The SHCS and the ZPHI have been approved by the ethics committee of the participating institutions (Kantonale Ethikkommission Bern, Ethikkommission des Kantons St. Gallen, Comite Departemental d’Ethique des Specialites Medicales et de Medicine Communataire et de Premier Recours, Kantonale Ethikkommission Zürich, Repubblica e Cantone Ticino - Comitato Ethico Cantonale, and Commission Cantonale d'Étique de la Recherche sur l'Être Humain, Ethikkommission Beider Basel for the SHCS, and Kantonale Ethikkommission Zürich for the ZPHI) and written informed consent had been obtained from all participants. Please see the supplementary note of Rusert et al. (2016) for further details on the cohorts.
Subcohorts
Our study cohort comprises a large proportion of individuals with highly similar background (white ethnicity and subtype B infection) and a second, smaller proportion with differential background (differential ethnicity, non-B infection; Fig. 1 and Table S2). Comparing our findings in the white/B subcohort with the full cohort provides powerful means to substantiate our conclusions as we see the same effects in both settings. As preferential recognition by plasma from different subtypes is unavoidable when analyzing binding responses to antigens of different subtypes, concordant data in both the white/B subcohort and the full cohort is particularly useful to exclude potential confounders.
In addition, we investigated subgroups of the white/B subcohort in Fig. 6 (see details of these subgroups in Fig. 5, A–C, and Table S2). To enable an unbiased evaluation, we restricted the analysis in Fig. 6 to the white/B subcohort and therein to individuals with chronic HIV-1 infection (3–10 yr untreated infection) to ensure comparable infection length in neutralizers and nonneutralizers. This was necessary as infection length is a driver of antibody development and bnAb activity in particular (Rusert et al., 2016). We previously demonstrated comparable bnAb frequency in individuals infected for >3 yr, but lower frequency in individuals infected for <3 yr (Rusert et al., 2016). We therefore excluded the latter individuals in the analysis in Fig. 6 to eliminate influences of infection length.
A further subgroup analysis of the white/B subcohort investigated early versus chronic HIV infection in Fig. 7. Data on the composition of these subgroups are shown in Fig. 5 C and Table S2.
Patient data
Both SHCS and ZPHI maintain a comprehensive, longitudinal, anonymous data collection of all participants, including extensive clinical and demographic data. The following data were used in the current study: longitudinal viral load and CD4 measurements, clinical history (antiretroviral drug history, infection length, and patient demographics), and pol nucleotide sequence data from genotypic antiretroviral drug resistance tests. The ethnicity (race) information is self-reported by the SHCS and ZPHI study participants at enrollment. Study nurses or study physicians use a structured interview questionnaire and participants are asked whether they belong to one of the following races/ethnicities: white, black, Hispano-American, or Asian. These categories cover the major ethnicities in Switzerland. Health care access is guaranteed for all races/ethnicities living in Switzerland and the same is true for participation in the SHCS and the ZPHI. We used pol data to estimate the viral diversity for two reasons: (1) we had access to pol data, as they were available from drug resistance tests. Full Env diversity determination for 4,281 individuals was not feasible. (2) pol data will give an unbiased estimate of diversity. Env diversity may be greater, but also has the chance to fluctuate more depending on whether the sampling time coincides with a dominant escape to the neutralization response or not. Please see the supplementary note of Rusert et al. (2016) for further details on how patient data were recorded. The following eight host, viral, and disease parameters were included in the analysis of the current study: viral load, CD4 count, infection length, virus diversity, gender, transmission mode, ethnicity, and viral pol subtype. The distributions of the parameters across the 4,281 patients analyzed in the current study are summarized in Table S2.
HIV-1 antigens
We selected 13 HIV-1 antigens covering structural Gag proteins (p17 and p24) and various Env antigens from different HIV-1 subtypes to establish HIV-binding antibody landscapes. Properties, sources, and details to production and purification of HIV-1 antigens are listed in Table S1 in (Liechti et al., 2018). We thank D. Burton (The Scripps Research Institute, La Jolla, CA), J.P. Moore (Cornell University, Ithaca, NY), R. Sanders (Academic Medical Center, Amsterdam, Netherlands), P.D. Kwong (National Institutes of Health Vaccine Research Center, Bethesda, MD), and J. Robinson (University of Zurich, Zurich, Switzerland) for providing proteins, peptides, or expression plasmids for this study.
We compared plasma antibody activity to various Env proteins and peptides: open Env conformations (JR-FL gp120 monomer [JR-FL gp120; Subtype B], BG505 gp140 monomer [BG505 gp140; Subtype A], V3 loop peptides of strains JR-FL, MN, and BG505 [V3 JR-FL, V3 MN, and V3 BG505]) with the closed, native-like trimer BG505 SOSIP.664 (BG505 trimer; Subtype A). To estimate CD4bs-directed responses, we probed plasma antibody activity to the V1-V3 loop truncated HxB2 derived core Env construct Resurfaced Stabilized Core 3 (RSC3) containing a preserved CD4bs and the RSC3Δ371Ι construct (RSC3Δ), which is not bound by most CD4bs antibodies (Wu et al., 2010). Within gp41, two MPER peptides, MPER-2/4 (encompassing the 2F5 and 4E10 bnAb epitopes) and MPER-2/4/10 (encompassing 2F5, 4E10, and 10E8 bnAb epitopes), and a truncated gp41 trimer construct lacking the MPER domain (gp41ΔMPER) were probed. All peptides were chemically biotinylated as indicated in Table S2. With the exception of p24 and gp41ΔMPER, all proteins were expressed with an avi-tag and enzymatically monobiotinylated (Avidity, LLC).
Profiling of the Swiss 4.5K cohort for HIV-1–binding antibodies using a customized multivariate multiplex bead assay
To measure the plasma IgG1-, IgG2-, and IgG3-binding antibody activity to the 13 selected Gag and Env HIV-1 antigens in the 4,281 included individuals, we used a high-throughput bead-based multiplexed immunoassay using the Luminex technology. We thank J. van Gils, T. Zborowski, and the Luminex Corporation (Hertogenbosch, Netherlands) for providing the FlexMap 3D instrument. The antigenicity of all 13 tested HIV-1 antigens in this Luminex-based binding assay has been previously established (Liechti et al., 2018). Importantly, the antigenicity of the BG505 trimer in this assay is identical to what was described previously with the BG505 trimer preferentially binding bnAbs and only allowing low level of non-/low-neutralizing V3 loop antibody binding compared with open Env versions (Yasmeen et al., 2014; Liechti et al., 2018).
Carboxylated MagPlex beads (Luminex) were covalently directly coupled with antigens (p24 and gp41ΔMPER) or coupled with Neutravidin (Thermo Fisher Scientific), followed by loading with biotinylated antigens (all others). For each antigen/analyte, a unique bead region was chosen to allow multiplexing of all 13 antigens. Coupling was done using a coupling kit (BioRad) according to the manufacturer’s instructions. In brief, 12.5 Mio beads were sonicated and thoroughly washed with the activation buffer provided by the coupling kit. The carboxyl groups on the beads were activated in 100 µl activation buffer containing sulfo-N-hydroxysulfosuccinimide (S-NHS; Thermo Fisher Scientific) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC; Thermo Fisher Scientific) at a concentration of 5 mg/ml. After 20 min of activation at room temperature, beads were thoroughly washed with activation buffer, and beads were coupled in 500 µl PBS containing the respective proteins (50 µg gp41ΔMPER, 62.5 µg p24, or 62.5 µg Neutravidin) for 2 h at room temperature. Beads were then washed thoroughly, blocked with PBS-BSA 1% for 30 min at room temperature, and, after extensive washing, stored in PBS-BSA 1% at a bead concentration of 20,000 beads/µl at 4°C.
Biotinylated proteins were coated on Neutravidin-coupled beads and subsequently blocked with biotin. Proteins and biotin were diluted in PBS at a concentration of 320 nM and coating and blocking were performed for 1 h at room temperature. Beads were extensively washed with PBS-BSA 1% and stored at a concentration of 10,000 beads/µl at 4°C and were used for up to 2 wk.
We established binding antibody profiles at fixed plasma dilutions of 1:100 (for all antigens and IgG subclasses) and 1:6,500 (IgG1 responses of highly reactive antigens p17, p24, gp120 JR-FL, and gp41ΔMPER).
The different bead regions carrying the different antigens were mixed at each experiment and incubated with patient plasma at dilution 1:100 and 1:6,500 in PBS-BSA 1% overnight at 4°C to allow plasma IgG binding to antigen-coated beads. Beads were washed with PBS-BSA 1%, distributed on 3 separate wells of 96-well plate, and IgG responses were detected with phycoerythrin-labeled secondary antibodies specific to isotypes IgG1, IgG2, and IgG3 diluted in PBS-BSA 1% at a concentration of 1 µg/ml (Southern Biotech). After extensive washing with PBS-BSA 1%, beads were analyzed with the FlexMap 3D instrument (Luminex). A minimum of 50 beads per antigen and plasma were acquired to guarantee accurate mean fluorescence intensity (MFI) values. Quality control and validation procedures for the FlexMap 3D instrument were done on each day of experiment according to manufacturer’s instructions, and monoclonal HIV-1–specific antibodies (PG09, 2F5, 19b, 2G12, 447-52D, and HIVIG) were analyzed with each experiment as assay quality control. IgG1 activity to p17, p24, gp120 JR-FL, and gp41ΔMPER is generally high and was thus analyzed at a 1:6,500 plasma dilution; all other antigens and IgG subclass responses were measured at a 1:100 plasma dilution to retrieve readout values in the dynamic range of the assay.
The binding reactivity of the 4,281 plasma samples was screened on 15 different days over a 2-mo period. Antibody reactivity to certain antigens showed only modest differences across the patient population (Fig. 1 C). In these cases, even small differences in measured binding intensities, caused, for instance, by day-to-day assay variability could potentially influence the analysis. We showed previously that influences of day-to-day variability can be prevented by transforming measured MFI values to per-day relative binding activities (Liechti et al., 2018). To retrieve the relative binding data, we considered for each antigen and IgG subclass combination the ordinal ranking of a plasma sample relative to all samples tested on the same day (ranging between 122 to 354 samples tested per day) and assigned a score of 0 to the lowest and 1 to the highest ranked sample and a linearly changing score to all other samples per day as described previously (Liechti et al., 2018; Table S1). This transformation was valid thanks to the high number of samples tested per day and allowed us to rule out a significant influence of single outliers. Importantly, the established intra- and interassay variability of the binding assay further allowed us to rely on single measurements of each plasma sample (Liechti et al., 2018). All postmeasurement analyses performed on the HIV-1 IgG–binding dataset except for the analysis reported in Fig. 9 used the relative binding data (Table S1).
Dissection of host, viral, and disease parameters that influence IgG binding
We used univariable and multivariable linear regression models to measure the influence of eight host, viral, and disease parameters on plasma IgG binding to the various HIV-1 antigens (using Python 2.7 and its library statsmodels0.8.0-3). Log10 viral load and CD4 level (both measured at time of sampling), viral pol diversity (Kouyos et al., 2011), and infection length were included as continuous variables. The remaining four factors were used as categorical variables and analyzed in relation to the reference category (sex: reference male; mode of transmission: reference men having sex with men [MSM]; ethnicity: reference white; HIV-1 subtype: reference subtype B). The multivariate analyses in Fig. 2 included all eight parameters. In the analyses in Fig. 3, the variables HIV-1 subtype and ethnicity were excluded, and only plasma samples of white individuals with subtype B infection (the white/B subcohort) were considered. In Fig. 4, the difference in plasma IgG binding between two IgG subclasses was compared for both the full cohort and the white/B subcohort. In this analysis, neutralization breadth is added as an additional categorical covariable. Patients were defined to have neutralization breadth if their plasma reached cross, broad, or elite neutralization activity score as determined in Rusert et al. (2016).
Predictive power of HIV-1 binding activities on HIV-1 neutralization breadth
Neutralization breadth scores for all tested plasma samples were available from the Swiss 4.5K Screen (Rusert et al., 2016) and are listed in Table S1. In brief, neutralization breadth scores were determined according to the neutralization of multiclade eight-virus panel using the Env pseudovirus luciferase reporter assay on TZM-bl cells (Rusert et al., 2016). Neutralization activity of plasma probed at a fixed dilution of 1:150 against a specific virus received a score of 0 when neutralization activity was <20%, a score of 1 when neutralization activity ranged between ≥20% and <50%, a score of 2 for neutralization between ≥50% and <80%, and a score of 3 for neutralization ≥80% (Rusert et al., 2016). The sum of scores against all 8 viruses reflects the cumulative potency and breadth neutralization score we used for further analysis. The maximum cumulative neutralization score a plasma could reach was 24. Scores of ≥15 were classified as elite neutralization, scores of 10–14 as broad neutralization, scores of 5–9 as cross neutralization, and scores <5 as no or weak neutralization activity.
To quantify the predictive power of a set of binding activities on HIV-1 neutralization activity, the set of all 4,281 plasma samples was partitioned into a training set (80%) and a test set (20%). Based on the training set, a univariable (if only one binding activity was considered as explanatory variable, as in Fig. 8, B and C) or multivariable (if multiple binding activities were considered as explanatory variables, as in Fig. 8, D–G) logistic regression model with breadth of neutralization response as the binary outcome variable was learned. We then used the learned model to rank the plasma samples of the test set and quantified the quality of the resulting ranking using the AUC of the ROC. In this analysis, a random predictor yields an AUC = 0.5, and a perfect predictor yields AUC = 1. We conducted three separate analyses for different choices of positives: at least cross neutralization (score > 4), at least broad neutralization (score > 9), and elite neutralization (score > 14). In each set-up, all remaining plasma samples were set as negatives. The reported mean AUC values are the result of 100 repetitions, each with fivefold cross-validation (i.e., partitioning of the set of all plasma samples into five equally sized sets and using each set four times as part of the training set and once as the test set). Uncertainties in the AUC values are not depicted graphically in Fig. 8 because the confidence intervals were negligibly small, limiting possibilities for visualization (see minimum and maximum AUC values in Table S4).
Profiling of the 105 broadest neutralizing plasma samples for HIV-1–binding antibody signatures
To allow a precise comparison of plasma IgG binding reactivity of the 105 broadest neutralizing plasma samples that we previously identified in the Swiss 4.5K Screen (Rusert et al., 2016), we subjected the 105 bnAb plasma to a separate assessment of HIV-1–binding activity in the multivariate multiplex bead assay using a wider range of plasma dilutions for IgG1 (1:400, 1:3,200, and 1:25,600) and a 1:400 plasma dilution for IgG2 and IgG3 to optimize measurements within the dynamic range of the respective responses. All samples were processed on a single day and measured on a Luminex FlexMap 3D reader. For each specific HIV-1 antigen, we considered an IgG1 dilution in the dynamic range of the assay. RSC3, RSC3Δ, MPER-2/4, and MPER-2/4/10 were measured in a 1:400 dilution, p24, and V3 MN in a 1:3,200 dilution, and p17, gp120 JR-FL, BG505 gp140, BG505 trimer, V3 JR-FL, V3 BG505, and gp41ΔMPER in a 1:25,600 dilution. The entire analysis was conducted in a single run excluding the possibility of assay-to-assay variations, as previously described (Liechti et al., 2018), and thus effective MFIs (Table S1), instead of relative binding data, could be used for the analysis reported in Fig. 9.
Predictive power of HIV-1–binding activities on bnAb specificity
To assess if specific HIV-1–binding antibody profiles predict bnAb specificity, we conducted multivariable linear regression analyses using detailed neutralization fingerprint data available for the top 105 bnAb plasmas and the HIV-1–binding antibody data available for the same patients (using Python 2.7 and its library statsmodels0.8.0-3). Details and results of the fingerprinting analysis of the 105 bnAb plasmas have been described (Rusert et al., 2016). In brief, half-maximum neutralization titer (NT50) of the 105 plasmas and 50% inhibitory concentrations (IC50) of bnAbs with known epitope specificities were determined against a 40 multiclade virus panel in the TZM-bl assay. bnAb specificity in plasma was then determined using a maximal Spearman–based prediction method (Rusert et al., 2016); that is, a plasma was assigned to the bnAb with the most similar neutralization fingerprint, where similarity was measured using the Spearman correlation of the IC50 values of the monoclonal antibodies and the inverse NT50 values of the plasmas. The retrieved Spearman correlation can be used as a proxy for the plasma bnAb specificity. Building on this data, we conducted in the current study multivariable linear regressions on the 105 bnAb plasmas with the maximal Spearman correlation of the neutralization fingerprint to a certain bnAb cluster (i.e., the plasma bnAb specificity) as the outcome variable and the binding activity to a certain antigen as the first explanatory variable (associations shown in Fig. 9 A). To avoid potential confounding effects (preferential reactivity with antigens and viruses of the same subtype), the viral pol subtype (B vs. non-B) was included as a second explanatory variable (associations shown in Fig. 9 B).
Blinding
Personnel who conducted the HIV-1–binding antibody analysis reported here and the previously reported neutralization screen (Rusert et al., 2016) had no information on patient demographics and neutralization activity at the time of analysis.
Statistical analysis
Statistical analyses were performed in Python 2.7 using the packages scipy.stats, statsmodels, and tsne as well as in R3.4.1 using its libraries circlize, car, and betareg.
Using the collected binding data, we used multivariable linear regression models to assess how individual factors influence HIV-1 IgG responses (Figs. 2 and 3), as well as how they influence the difference in responses between two IgG subclasses (Fig. 4). Although only multivariable analyses have the power to disentangle confounded effects, results from univariable analysis are additionally provided for each analysis, as divergent results in univariable and multivariable analyses best highlight which effects are most affected by other variables and can only be reliably assessed by multivariable tests. This provides important information, which needs to be considered in studies that, because of limited sample numbers, can only use univariable testing. We tested the appropriateness of the obtained linear models. As the relative binding activities are close to uniformly distributed in [0,1], outliers are not an issue. The ncv-test (R library car), which tests for heteroscedasticity, revealed 26 out of 156 investigated models with P < 0.05; only two models showed significant heteroscedasticity after correcting for multiple testing. Given the large number of observations and the moderate p-values, heteroscedasticity is unlikely to have affected our results. Lastly, as a sensitivity analysis to Figs. 2 and 3, we investigated additional, more complicated β regression models (using a linear transformation of the response variables from [0,1] into (0,1); Smithson and Verkuilen, 2006) that take the distribution of the relative binding activities into account. These models revealed very similar associations as reported in Figs. 2 and 3 (not depicted). We thus opted to report the results from the more frequently used and more easily interpretable linear regression analyses.
In Figs. 6 and 7, Spearman correlations were calculated for each pair of relative binding activities (respective parts A and B) and compared between the two subpopulations under investigation (respective parts C and D). To test for significant differences in pairwise Spearman correlations (part C), the two subpopulations were randomly reshuffled 10,000 times, keeping only the respective sizes fixed. Each time, the differences in Spearman correlations between the reshuffled subpopulations were recorded and finally compared with the observed differences found in the true subpopulations. If there was no true difference in correlation between the two subpopulations, ∼5,000 of the reshuffled correlations were expected to be higher than the observed correlation and 5,000 lower. We thus calculated the p-value of an observed correlation as
where m is the number of reshuffled correlations higher than the observed correlation. To test whether two antibody response landscapes exhibit significantly different “between IgG subclass” Spearman correlations, we compared correlations between same antigens (part D) using the paired Wilcoxon test. The dimensionality reduction method t-SNE was used to display the plasma samples in two dimensions, taking into account their relative IgG1 BG505 trimer, IgG2 BG505 trimer, IgG1 MPER-2/4, and IgG3 MPER-2/4 binding activities (Fig. 10). As a result of the large cohort size, we used the well-established Barnes-Hut-SNE approximation (with 1,000 iterations and parameters theta = 0.5 and perplexity = 200) instead of the exact t-SNE method (van der Maaten, 2013).
Multiple testing
All reported statistical analyses are of explorative, descriptive nature. We therefore opted, by default, not to formally adjust for multiple testing because false positives are less of a problem in explorative studies than false negatives. In addition, owing to the large size of our cohort, almost all the associations we focus on exhibit a very low p-value, much lower than a Bonferroni corrected p-value (pBonferroni = 0.05/3 IgG isotypes/13 antigens/8–11 predictors > 10−4) as can be seen in Figs. 2 and 3. Levels of significance for Bonferroni-corrected p-values are shown in Figs. 2 and 3 and allow assessment of multiple testing corrected values.
In Figs. 6 and 7, we used a Benjamini-Hochberg correction with a false discovery rate of 10% to correct for multiple testing. The analysis reported in Fig. 9 was performed solely to test the association of bnAb specificities and binding activities for two bnAb types, CD4bs and MPER, and specific antigens (RSC3/RSC3Δ and MPER peptides, respectively). We reported the results for the additional antigens and bnAb specificities only for completeness, but did not draw any conclusions because the outcome cannot be interpreted appropriately without having bnAb type-specific antigens.
Online supplemental material
Table S1 contains all raw data (patient characteristics, relative binding activities, the effective measured binding responses [MFIs], and bnAb specificities for the 105 individuals with highest bnAb activity). Table S2 shows the characteristics of the various patient subpopulations analyzed in this study. Table S3 contains the detailed regression results of the analyses shown in Figs. 2 and 3. Table S4 contains the mean AUC values for univariable and multivariable prediction models of neutralization breadth shown in Fig. 8. Table S5 contains the detailed regression results of the analyses shown in Fig. 9.
Supplementary Material
Acknowledgments
We thank the patients participating in the ZPHI and the SHCS and their physicians and study nurses for patient care and D. Perraudin and M. Minichiello for administrative assistance. We are very grateful to J. van Gils, T. Zborowski, and the Luminex Corporation for providing the FlexMap 3D instrument for the HIV IgG measurements.
Financial support for this study has been provided by the Swiss National Science Foundation (SNF; grants 314730_152663 and 314730_172790 to A. Trkola and grant 324730_159868 to H.F. Günthard), the Clinical Priority Research Program of the University of Zurich (Viral infectious diseases: Zurich Primary HIV Infection Study to H.F. Günthard and A. Trkola), the Yvonne-Jacob Foundation (to H.F. Günthard) the Swiss Vaccine Research Institute (to A. Trkola, H.F. Günthard, and R.D. Kouyos) and the SystemsX.ch grant AntibodyX (to A. Trkola). R.D. Kouyos was supported by the SNF (grants PZ00P3-142411 and BSSGI0_155851). This study has been cofinanced within the framework of the Swiss HIV Cohort Study (SHCS), supported by the SNF (grants 33CS30_148522 to H.F. Günthard), by the small nested SHCS project 744 (to A. Trkola), and by the SHCS Research Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The SHCS data are collected by the five Swiss University Hospitals, two Cantonal Hospitals, 15 affiliated hospitals, and 36 private physicians (listed in http://www.shcs.ch/180-health-care-providers).
The authors declare no competing financial interests.
Author Contributions: A. Trkola, H.F. Günthard, and R.D. Kouyos jointly directed this work. C. Kadelka, T. Liechti, H. Ebner, P. Rusert, R.D. Kouyos, H.F. Günthard, and A. Trkola conceived and designed the study and analyzed data. T. Liechti, H. Ebner, and P. Rusert designed and performed experiments and analyzed data. J. Weber, T. Uhr, H. Kuster, S. Yerly, V. Aubert, T. Klimkait, J. Böni, B. Misselwitz, M. Schanz, M. Huber, N. Friedrich, and E. Stiegeler conducted experiments and analyzed data. D.L. Braun, M. Cavassini, E. Bernasconi, M. Hoffmann, A. Calmy, M. Battegay, A. Rauch, A.U. Scherrer, H.F. Günthard, and the members of the Swiss HIV Cohort Study conceived and managed the SHCS, and ZPHI cohorts collected and contributed patient samples and clinical data. C. Kadelka, T. Liechti, R.D. Kouyos, M. Schanz, N. Friedrich, H.F. Günthard, and A. Trkola wrote the manuscript, which all coauthors commented on.
References
- Ackerman M.E., Mikhailova A., Brown E.P., Dowell K.G., Walker B.D., Bailey-Kellogg C., Suscovich T.J., and Alter G.. 2016. Polyfunctional HIV-Specific Antibody Responses Are Associated with Spontaneous HIV Control. PLoS Pathog. 12:e1005315 10.1371/journal.ppat.1005315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman M.E., Barouch D.H., and Alter G.. 2017. Systems serology for evaluation of HIV vaccine trials. Immunol. Rev. 275:262–270. 10.1111/imr.12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K., Klasse P.J., Sanders R.W., Pereyra F., Michael E., Lu M., Walker B.D., and Moore J.P.. 2010. IgG subclass profiles in infected HIV type 1 controllers and chronic progressors and in uninfected recipients of Env vaccines. AIDS Res. Hum. Retroviruses. 26:445–458. 10.1089/aid.2009.0223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhiman J.N., Anthony C., Doria-Rose N.A., Karimanzira O., Schramm C.A., Khoza T., Kitchin D., Botha G., Gorman J., Garrett N.J., et al. . 2015. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat. Med. 21:1332–1336. 10.1038/nm.3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley J.M., Klasse P.J., Cao Y., Jones I., Markowitz M., Ho D.D., and Moore J.P.. 1997. Differential regulation of the antibody responses to Gag and Env proteins of human immunodeficiency virus type 1. J. Virol. 71:2799–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongertz V., Guimarães M.L., Soares-da-Costa M.F., Veloso V.G., Bastos F.I., Szwarcwald C.L., Derrico M., Telles P.R., Pilloto J.H., João Filho E.C., and Morgado M.G.. The HEC/FIOCRUZ AIDS Clinical Research Group . 1999. Anti-HIV-1 seroreactivity and HIV transmission route[R1]. J. Clin. Virol. 12:27–36. 10.1016/S0928-0197(98)00068-3 [DOI] [PubMed] [Google Scholar]
- Borrow P., and Moody M.A.. 2017. Immunologic characteristics of HIV-infected individuals who make broadly neutralizing antibodies. Immunol. Rev. 275:62–78. 10.1111/imr.12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broliden P.A., Morfeldt-Månsson L., Rosen J., Jondal M., and Wahren B.. 1989. Fine specificity of IgG subclass response to group antigens in HIV-1-infected patients. Clin. Exp. Immunol. 76:216–221. [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., and Hangartner L.. 2016. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu. Rev. Immunol. 34:635–659. 10.1146/annurev-immunol-041015-055515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey M., Klein F., and Nussenzweig M.C.. 2016. Broadly Neutralizing Antibodies for HIV-1 Prevention or Immunotherapy. N. Engl. J. Med. 375:2019–2021. 10.1056/NEJMp1613362 [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhang J., Hwang K.K., Bouton-Verville H., Xia S.M., Newman A., Ouyang Y.B., Haynes B.F., and Verkoczy L.. 2013. Common tolerance mechanisms, but distinct cross-reactivities associated with gp41 and lipids, limit production of HIV-1 broad neutralizing antibodies 2F5 and 4E10. J. Immunol. 191:1260–1275. 10.4049/jimmunol.1300770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung A.W., Kumar M.P., Arnold K.B., Yu W.H., Schoen M.K., Dunphy L.J., Suscovich T.J., Frahm N., Linde C., Mahan A.E., et al. . 2015. Dissecting Polyclonal Vaccine-Induced Humoral Immunity against HIV Using Systems Serology. Cell. 163:988–998. 10.1016/j.cell.2015.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez V., Odem-Davis K., McClelland R.S., Jaoko W., and Overbaugh J.. 2012. HIV-1 superinfection in women broadens and strengthens the neutralizing antibody response. PLoS Pathog. 8:e1002611 10.1371/journal.ppat.1002611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N.A., Klein R.M., Daniels M.G., O’Dell S., Nason M., Lapedes A., Bhattacharya T., Migueles S.A., Wyatt R.T., Korber B.T., et al. . 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631–1636. 10.1128/JVI.01482-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N.A., Schramm C.A., Gorman J., Moore P.L., Bhiman J.N., DeKosky B.J., Ernandes M.J., Georgiev I.S., Kim H.J., Pancera M., et al. NISC Comparative Sequencing Program . 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 509:55–62. 10.1038/nature13036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugast A.S., Arnold K., Lofano G., Moore S., Hoffner M., Simek M., Poignard P., Seaman M., Suscovich T.J., Pereyra F., et al. . 2017. Virus-driven Inflammation Is Associated With the Development of bNAbs in Spontaneous Controllers of HIV. Clin. Infect. Dis. 64:1098–1104. 10.1093/cid/cix057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolano A., Dosenovic P., and Nussenzweig M.C.. 2017. Progress toward active or passive HIV-1 vaccination. J. Exp. Med. 214:3–16. 10.1084/jem.20161765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelman F.D., Katona I.M., Mosmann T.R., and Coffman R.L.. 1988. IFN-gamma regulates the isotypes of Ig secreted during in vivo humoral immune responses. J. Immunol. 140:1022–1027. [PubMed] [Google Scholar]
- Fish E.N. 2008. The X-files in immunity: sex-based differences predispose immune responses. Nat. Rev. Immunol. 8:737–744. 10.1038/nri2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French M.A., Tanaskovic S., Law M.G., Lim A., Fernandez S., Ward L.D., Kelleher A.D., and Emery S.. 2010. Vaccine-induced IgG2 anti-HIV p24 is associated with control of HIV in patients with a ‘high-affinity’ FcgammaRIIa genotype. AIDS. 24:1983–1990. 10.1097/QAD.0b013e32833c1ce0 [DOI] [PubMed] [Google Scholar]
- French M.A., Center R.J., Wilson K.M., Fleyfel I., Fernandez S., Schorcht A., Stratov I., Kramski M., Kent S.J., and Kelleher A.D.. 2013. Isotype-switched immunoglobulin G antibodies to HIV Gag proteins may provide alternative or additional immune responses to ‘protective’ human leukocyte antigen-B alleles in HIV controllers. AIDS. 27:519–528. 10.1097/QAD.0b013e32835cb720 [DOI] [PubMed] [Google Scholar]
- Goo L., Chohan V., Nduati R., and Overbaugh J.. 2014. Early development of broadly neutralizing antibodies in HIV-1-infected infants. Nat. Med. 20:655–658. 10.1038/nm.3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray E.S., Madiga M.C., Hermanus T., Moore P.L., Wibmer C.K., Tumba N.L., Werner L., Mlisana K., Sibeko S., Williamson C., et al. CAPRISA002 Study Team . 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85:4828–4840. 10.1128/JVI.00198-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen S., and Altfeld M.. 2016. The X awakens: multifactorial ramifications of sex-specific differences in HIV-1 infection. J. Virus Erad. 2:78–81. [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C., Carnathan D.G., Torrents de la Peña A., Pauthner M., Briney B., Reiss S.M., Wood J.S., Kaushik K., van Gils M.J., Rosales S.L., et al. . 2016. Direct Probing of Germinal Center Responses Reveals Immunological Features and Bottlenecks for Neutralizing Antibody Responses to HIV Env Trimer. Cell Reports. 17:2195–2209. 10.1016/j.celrep.2016.10.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F., Fleming J., St Clair E.W., Katinger H., Stiegler G., Kunert R., Robinson J., Scearce R.M., Plonk K., Staats H.F., et al. . 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 308:1906–1908. 10.1126/science.1111781 [DOI] [PubMed] [Google Scholar]
- Haynes B.F., Shaw G.M., Korber B., Kelsoe G., Sodroski J., Hahn B.H., Borrow P., and McMichael A.J.. 2016. HIV-Host Interactions: Implications for Vaccine Design. Cell Host Microbe. 19:292–303. 10.1016/j.chom.2016.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horns F., Vollmers C., Croote D., Mackey S.F., Swan G.E., Dekker C.L., Davis M.M., and Quake S.R.. 2016. Lineage tracing of human B cells reveals the in vivo landscape of human antibody class switching. eLife. 5:e16578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraber P., Seaman M.S., Bailer R.T., Mascola J.R., Montefiori D.C., and Korber B.T.. 2014. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 28:163–169. 10.1097/QAD.0000000000000106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ofek G., Laub L., Louder M.K., Doria-Rose N.A., Longo N.S., Imamichi H., Bailer R.T., Chakrabarti B., Sharma S.K., et al. . 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 491:406–412. 10.1038/nature11544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J.G., Kulp D.W., Havenar-Daughton C., Sarkar A., Briney B., Sok D., Sesterhenn F., Ereño-Orbea J., Kalyuzhniy O., Deresa I., et al. . 2016. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 351:1458–1463. 10.1126/science.aad9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y., Noma T., and Yata J.. 1994. Regulation of human IgG subclass production by cytokines. IFN-gamma and IL-6 act antagonistically in the induction of human IgG1 but additively in the induction of IgG2. J. Immunol. 153:4948–4958. [PubMed] [Google Scholar]
- Kelsoe G., and Haynes B.F.. 2017. Host controls of HIV broadly neutralizing antibody development. Immunol. Rev. 275:79–88. 10.1111/imr.12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Excler J.L., and Michael N.L.. 2015. Lessons from the RV144 Thai phase III HIV-1 vaccine trial and the search for correlates of protection. Annu. Rev. Med. 66:423–437. 10.1146/annurev-med-052912-123749 [DOI] [PubMed] [Google Scholar]
- Klasse P.J., Blomberg J., and Pipkorn R.. 1990. Differential IgG subclass responses to epitopes in transmembrane protein of HIV-1. Viral Immunol. 3:89–98. 10.1089/vim.1990.3.89 [DOI] [PubMed] [Google Scholar]
- Knox J.J., Buggert M., Kardava L., Seaton K.E., Eller M.A., Canaday D.H., Robb M.L., Ostrowski M.A., Deeks S.G., Slifka M.K., et al. . 2017. T-bet+ B cells are induced by human viral infections and dominate the HIV gp140 response. JCI Insight. 2:e92943 10.1172/jci.insight.92943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyos R.D., von Wyl V., Yerly S., Böni J., Rieder P., Joos B., Taffé P., Shah C., Bürgisser P., Klimkait T., et al. Swiss HIV Cohort Study . 2011. Ambiguous nucleotide calls from population-based sequencing of HIV-1 are a marker for viral diversity and the age of infection. Clin. Infect. Dis. 52:532–539. 10.1093/cid/ciq164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J.I., Licht A.F., Dugast A.S., Suscovich T., Choi I., Bailey-Kellogg C., Alter G., and Ackerman M.E.. 2014. Divergent antibody subclass and specificity profiles but not protective HLA-B alleles are associated with variable antibody effector function among HIV-1 controllers. J. Virol. 88:2799–2809. 10.1128/JVI.03130-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal R.B., Heiba I.M., Dhawan R.R., Smith E.S., and Perine P.L.. 1991. IgG subclass responses to human immunodeficiency virus-1 antigens: lack of IgG2 response to gp41 correlates with clinical manifestation of disease. Clin. Immunol. Immunopathol. 58:267–277. 10.1016/0090-1229(91)90141-V [DOI] [PubMed] [Google Scholar]
- Landais E., Huang X., Havenar-Daughton C., Murrell B., Price M.A., Wickramasinghe L., Ramos A., Bian C.B., Simek M., Allen S., et al. . 2016. Broadly Neutralizing Antibody Responses in a Large Longitudinal Sub-Saharan HIV Primary Infection Cohort. PLoS Pathog. 12:e1005369 10.1371/journal.ppat.1005369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti T., Kadelka C., Ebner H., Friedrich N., Kouyos R.D., Günthard H.F., and Trkola A.. 2018. Development of a high-throughput bead based assay system to measure HIV-1 specific immune signatures in clinical samples. J. Immunol. Methods. 454:48–58. 10.1016/j.jim.2017.12.003 [DOI] [PubMed] [Google Scholar]
- Martinez V., Costagliola D., Bonduelle O., N’go N., Schnuriger A., Théodorou I., Clauvel J.P., Sicard D., Agut H., Debré P., et al. Asymptomatiques à Long Terme Study Group . 2005. Combination of HIV-1-specific CD4 Th1 cell responses and IgG2 antibodies is the best predictor for persistence of long-term nonprogression. J. Infect. Dis. 191:2053–2063. 10.1086/430320 [DOI] [PubMed] [Google Scholar]
- Mellors J.W., Muñoz A., Giorgi J.V., Margolick J.B., Tassoni C.J., Gupta P., Kingsley L.A., Todd J.A., Saah A.J., Detels R., et al. . 1997. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann. Intern. Med. 126:946–954. 10.7326/0003-4819-126-12-199706150-00003 [DOI] [PubMed] [Google Scholar]
- Moody M.A., Pedroza-Pacheco I., Vandergrift N.A., Chui C., Lloyd K.E., Parks R., Soderberg K.A., Ogbe A.T., Cohen M.S., Liao H.-X., et al. . 2016. Immune perturbations in HIV-1-infected individuals who make broadly neutralizing antibodies. Sci. Immunol. 1:aag0851 10.1126/sciimmunol.aag0851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.L., and Williamson C.. 2016. Approaches to the induction of HIV broadly neutralizing antibodies. Curr. Opin. HIV AIDS. 11:569–575. 10.1097/COH.0000000000000317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T., Steindl F., Purtscher M., Trkola A., Klima A., Himmler G., Rüker F., and Katinger H.. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo-Giang-Huong N., Candotti D., Goubar A., Autran B., Maynart M., Sicard D., Clauvel J.P., Agut H., Costagliola D., and Rouzioux C.. French Asymptomatic Long-Term Study Group . 2001. HIV type 1-specific IgG2 antibodies: markers of helper T cell type 1 response and prognostic marker of long-term nonprogression. AIDS Res. Hum. Retroviruses. 17:1435–1446. 10.1089/088922201753197105 [DOI] [PubMed] [Google Scholar]
- Pegu A., Hessell A.J., Mascola J.R., and Haigwood N.L.. 2017. Use of broadly neutralizing antibodies for HIV-1 prevention. Immunol. Rev. 275:296–312. 10.1111/imr.12511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piantadosi A., Panteleeff D., Blish C.A., Baeten J.M., Jaoko W., McClelland R.S., and Overbaugh J.. 2009. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J. Virol. 83:10269–10274. 10.1128/JVI.01149-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder P., Joos B., Scherrer A.U., Kuster H., Braun D., Grube C., Niederöst B., Leemann C., Gianella S., Metzner K.J., et al. . 2011. Characterization of human immunodeficiency virus type 1 (HIV-1) diversity and tropism in 145 patients with primary HIV-1 infection. Clin. Infect. Dis. 53:1271–1279. 10.1093/cid/cir725 [DOI] [PubMed] [Google Scholar]
- Rusert P., Kouyos R.D., Kadelka C., Ebner H., Schanz M., Huber M., Braun D.L., Hozé N., Scherrer A., Magnus C., et al. Swiss HIV Cohort Study . 2016. Determinants of HIV-1 broadly neutralizing antibody induction. Nat. Med. 22:1260–1267. 10.1038/nm.4187 [DOI] [PubMed] [Google Scholar]
- Sanders R.W., van Gils M.J., Derking R., Sok D., Ketas T.J., Burger J.A., Ozorowski G., Cupo A., Simonich C., Goo L., et al. . 2015. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 349:aac4223 10.1126/science.aac4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather D.N., Armann J., Ching L.K., Mavrantoni A., Sellhorn G., Caldwell Z., Yu X., Wood B., Self S., Kalams S., and Stamatatos L.. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769. 10.1128/JVI.02036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau Q.J., and Moore J.P.. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185–196. 10.1084/jem.182.1.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeni-Affolter F., Ledergerber B., Rickenbach M., Rudin C., Günthard H.F., Telenti A., Furrer H., Yerly S., and Francioli P.. Swiss HIV Cohort Study . 2010. Cohort profile: the Swiss HIV Cohort study. Int. J. Epidemiol. 39:1179–1189. 10.1093/ije/dyp321 [DOI] [PubMed] [Google Scholar]
- Smithson M., and Verkuilen J.. 2006. A better lemon squeezer? Maximum-likelihood regression with beta-distributed dependent variables. Psychol. Methods. 11:54–71. 10.1037/1082-989X.11.1.54 [DOI] [PubMed] [Google Scholar]
- Stamatatos L., Pancera M., and McGuire A.T.. 2017. Germline-targeting immunogens. Immunol. Rev. 275:203–216. 10.1111/imr.12483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J. 1996. Immunoglobulin class switching. Curr. Opin. Immunol. 8:199–205. 10.1016/S0952-7915(96)80058-6 [DOI] [PubMed] [Google Scholar]
- Sterling T.R., Vlahov D., Astemborski J., Hoover D.R., Margolick J.B., and Quinn T.C.. 2001. Initial plasma HIV-1 RNA levels and progression to AIDS in women and men. N. Engl. J. Med. 344:720–725. 10.1056/NEJM200103083441003 [DOI] [PubMed] [Google Scholar]
- Stiegler G., Kunert R., Purtscher M., Wolbank S., Voglauer R., Steindl F., and Katinger H.. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses. 17:1757–1765. 10.1089/08892220152741450 [DOI] [PubMed] [Google Scholar]
- Thomas H.I., Wilson S., O’Toole C.M., Lister C.M., Saeed A.M., Watkins R.P., and Morgan-Capner P.. 1996. Differential maturation of avidity of IgG antibodies to gp41, p24 and p17 following infection with HIV-1. Clin. Exp. Immunol. 103:185–191. 10.1046/j.1365-2249.1996.951642.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras G.D., and Haynes B.F.. 2009. HIV-1-specific antibody responses during acute and chronic HIV-1 infection. Curr. Opin. HIV AIDS. 4:373–379. 10.1097/COH.0b013e32832f00c0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A., Kuster H., Leemann C., Oxenius A., Fagard C., Furrer H., Battegay M., Vernazza P., Bernasconi E., Weber R., et al. Swiss HIV Cohort Study . 2004. Humoral immunity to HIV-1: kinetics of antibody responses in chronic infection reflects capacity of immune system to improve viral set point. Blood. 104:1784–1792. 10.1182/blood-2004-01-0251 [DOI] [PubMed] [Google Scholar]
- van der Maaten L.2013. Barnes-Hut-SNE. arXiv https://arxiv.org/abs/1301.3342
- Vazquez M.I., Catalan-Dibene J., and Zlotnik A.. 2015. B cells responses and cytokine production are regulated by their immune microenvironment. Cytokine. 74:318–326. 10.1016/j.cyto.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voltersvik P., Albrektsen G., Ulvestad E., Dyrhol-Riise A.M., Sørensen B., and Asjö B.. 2003. Changes in immunoglobulin isotypes and immunoglobulin G (IgG) subclasses during highly active antiretroviral therapy: anti-p24 IgG1 closely parallels the biphasic decline in plasma viremia. J. Acquir. Immune Defic. Syndr. 34:358–367. 10.1097/00126334-200312010-00002 [DOI] [PubMed] [Google Scholar]
- Ward A.B., and Wilson I.A.. 2017. The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol. Rev. 275:21–32. 10.1111/imr.12507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Yang Z.Y., Li Y., Hogerkorp C.M., Schief W.R., Seaman M.S., Zhou T., Schmidt S.D., Wu L., Xu L., et al. . 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 329:856–861. 10.1126/science.1187659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.L., Kouyos R., Scherrer A.U., Böni J., Shah C., Yerly S., Klimkait T., Aubert V., Furrer H., Battegay M., et al. Swiss HIV Cohort Study . 2015. Assessing the Paradox Between Transmitted and Acquired HIV Type 1 Drug Resistance Mutations in the Swiss HIV Cohort Study From 1998 to 2012. J. Infect. Dis. 212:28–38. 10.1093/infdis/jiv012 [DOI] [PubMed] [Google Scholar]
- Yasmeen A., Ringe R., Derking R., Cupo A., Julien J.P., Burton D.R., Ward A.B., Wilson I.A., Sanders R.W., Moore J.P., and Klasse P.J.. 2014. Differential binding of neutralizing and non-neutralizing antibodies to native-like soluble HIV-1 Env trimers, uncleaved Env proteins, and monomeric subunits. Retrovirology. 11:41 10.1186/1742-4690-11-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates N.L., Liao H.X., Fong Y., deCamp A., Vandergrift N.A., Williams W.T., Alam S.M., Ferrari G., Yang Z.Y., Seaton K.E., et al. . 2014. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci. Transl. Med. 6:228ra39 10.1126/scitranslmed.3007730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong M.K., Cameron P.U., Spelman T., Elliott J.H., Fairley C.K., Boyle J., Miyamasu M., and Lewin S.R.. 2016. Quantifying Adaptive and Innate Immune Responses in HIV-Infected Participants Using a Novel High Throughput Assay. PLoS One. 11:e0166549 10.1371/journal.pone.0166549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H., and Casali P.. 2015. Epigenetics of Peripheral B-Cell Differentiation and the Antibody Response. Front. Immunol. 6:631 10.3389/fimmu.2015.00631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Verkoczy L., Wiehe K., Munir Alam S., Nicely N.I., Santra S., Bradley T., Pemble C.W. IV, Zhang J., Gao F., et al. . 2016. Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages. Sci. Transl. Med. 8:336ra62 10.1126/scitranslmed.aaf0618 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.