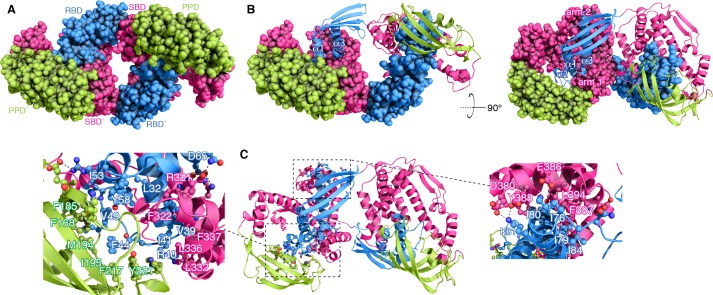
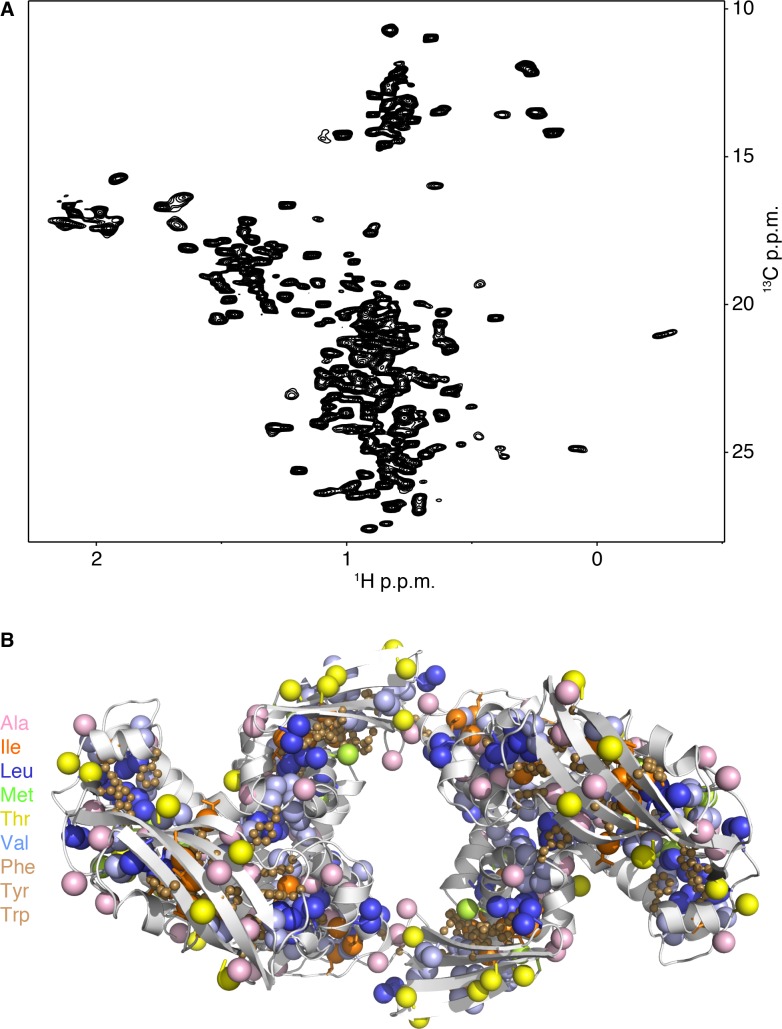
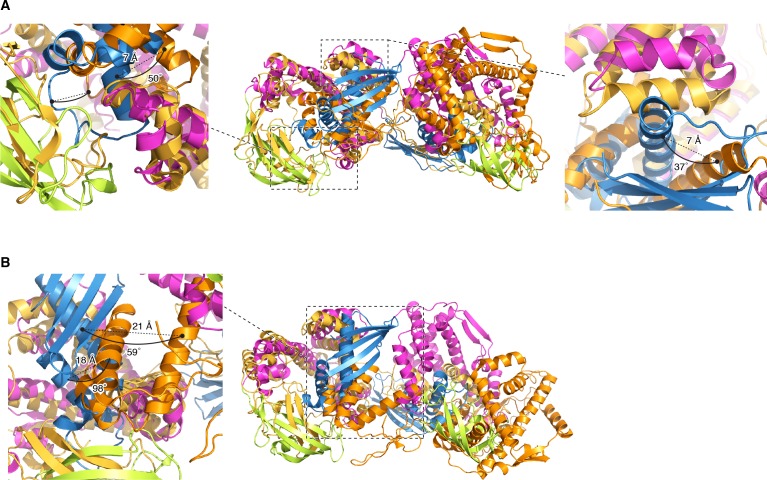
The structure of TF dimer superimposed with previously reported PRE-based docking models (
Morgado et al., 2017); conformer 1 [PDB code: 5OWI] (
A), and conformer 2 [PDB code: 5OWJ] (
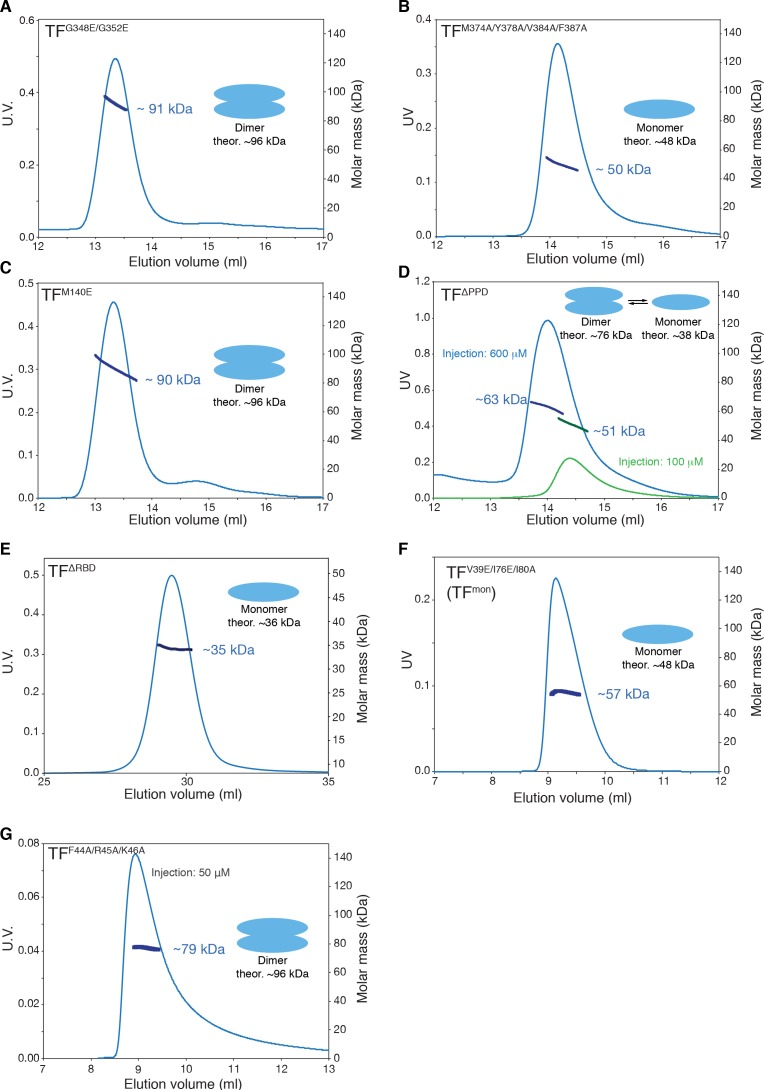
B). PPD, SBD, and RBD in the structure of TF dimer are shown in green, pink, and blue, respectively. The docking model is shown in orange and gold. The coordinates are superimposed on the backbone heavy atoms of SBD. Differences in rotation and translation of the helices in RBD between the previously reported PRE-based docking model and the current dimer structure are indicated. Most of the contacts, which are seen in the current structure of the TF dimer and were validated by mutagenesis and chemical shift perturbation mapping, are not present in the PRE-based models. In the PRE-derived conformer 1, RBD makes contacts with the arm 1 and the PPD of the other subunit, which were also seen in the current dimer TF structure. However, in the PRE-derived conformer 1 the RBD makes no significant contacts with the arm 2, which is not consistent with the significant effect that mutations in the arm 2 have on the dimerization as shown by SEC-MALS (
Figure 2—figure supplement 2B). The PRE-derived conformer 2 has RBD snagged on the tips of the arm 1 and arm 2, as well as on the edge of PPD. A very small overlap between the substrate-binding sites and the dimer interface is seen in the PRE-derived conformer 2. In contrast, in the current structure of TF dimer, RBD is buried inside the cradle formed by SBD and PPD of the other subunit, thus explaining why the TF dimer dissociates upon binding to the substrate protein (
Figure 1—figure supplement 1A,E and F). In addition to the significant differences in the overall domain orientation between the PRE-derived models and the current TF structure, the RBD structure itself appears to be loosely packed in the PRE-derived models.