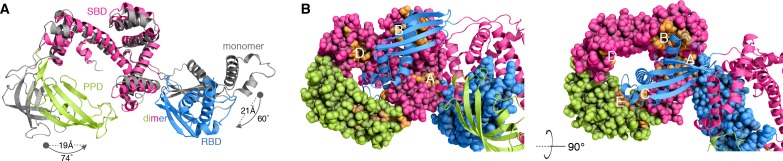
Figure 3. Conformational changes of TF upon dimerization.
(A) The structure of one subunit in the TF dimer (colored as in Figure 1A) and the crystal structure of monomeric TF (colored grey) [Protein Data Bank (PDB) code: 1W26] are superimposed for SBD. The changes in rotation and translation of the RBD and PPD between the monomer and the dimer are indicated. (B) View of the structure of dimeric TF highlighting the positioning of the substrate-binding sites (colored orange). The five main substrate-binding sites are labeled A, B, C, D, and E.