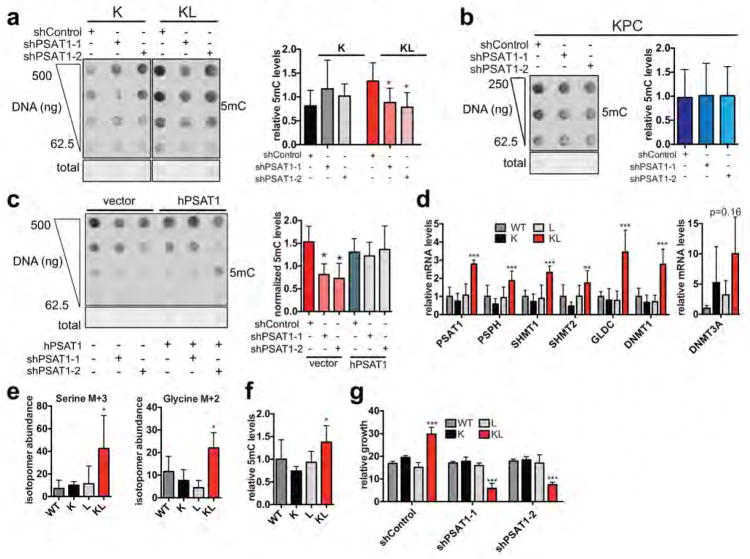
Extended Data Figure 6. Serine pathway activity sustains DNA methylation in KL cells.
a, Dot blot of DNA isolated from K or KL cells transduced with shControl, shPSAT1-1 or shPSAT1-2 probed with anti-5mC antibody. Total DNA was visualized by methylene blue staining. Graph shows quantitated signal normalized to total DNA as measured by methylene blue staining (K cells, n=4, KL cells, n=8, independent replicates). b, Dot blot of DNA isolated from KPC cells transduced with shControl, shPSAT1-1 or shPSAT1-2 probed with anti-5mC antibody. The graph shows quantitated signal normalized to total DNA (n=4, independent replicates). c, Dot blot of DNA probed with anti-5mC antibody. DNA was isolated from KL cells first transduced with vector or human PSAT1 then transduced with shControl, shPSAT1-1/2. Graph shows quantitated signal normalized to total DNA as measured by methylene blue staining (n=3, independent replicates). d, Expression of serine pathway genes and DNMTs genes in WT, K, L or KL cells by qRT-PCR. Data are normalized to 18S and expressed as relative to K (n=6, pooled data from two experiments). e, Plots of isotopomer abundance of 13C-U-Glucose-derived M+3 serine and M+2 glycine 6 hrs after additoin of 13C-U-Glucose (n=6 independent biological replicates). f, Quantitated DNA dot blot signal of DNA isolated from WT, K, L or KL cells probed with anti-5-methyl-cytosine antibody (5mC) normalized to total DNA as measured by methylene blue staining (n=4, independent replicates). g, Five-day growth of WT, K, L or KL cells transduced with shControl or two hairpins against PSAT1. Data are expressed as relative to day 0 (n=4, pooled from two experiments). For all panels, error bars are standard deviation unless otherwise stated and statistical significance is indicated as follows: *P<0.05, **P<0.01, ***P<0.001.