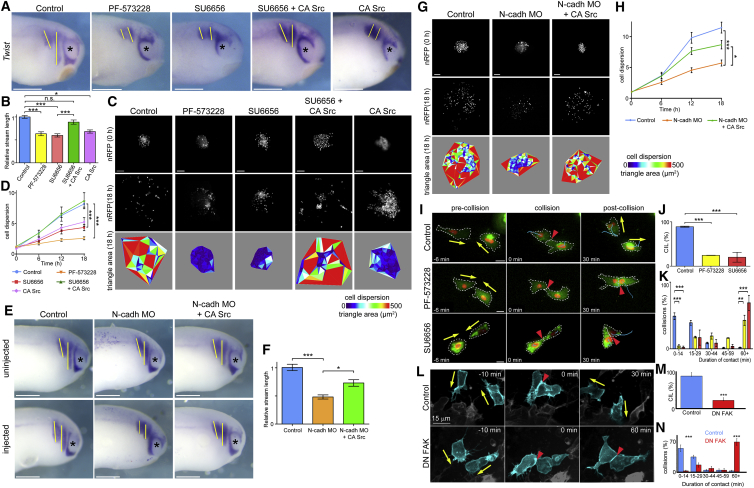
Figure 6.
FAK-Src Signaling Required for NC Dispersion, Migration, and CIL
(A) Migrating NC labeled with twist in embryos at stage 24, after the indicated treatments. Yellow lines, distance of NC migration; asterisks, eye. Scale bar, 500 μm.
(B) Length of second NC stream relative to mean control of each experiment. Control n = 101, PF-573228 n = 47, SU6656 n = 46, SU6656 + CA Src n = 35, CA Src n = 7 embryos.
(C) Initial frame (0 hr) and last frame (18 hr) from movies of explanted NC cell clusters expressing nuclear RFP, after the indicated treatments. Dispersion analysis based on Delaunay triangles color coded for area size. Scale bar: 100 μm.
(D) Normalized mean triangle areas over time. Control n = 52, PF-573228 n = 28, SU6656 n = 32, SU6656 + Src Y527F n = 32, CA Src n = 10 explants.
(E) Migrating NC labeled with twist in embryos at stage 24, after the indicated treatments. Yellow lines, distance of NC migration; asterisks, eye. Scale bar, 500 μm.
(F) Length of second NC stream migration of injected side relative to uninjected side. n = 33 embryos for control, n = 30 embryos for N-cadherin MO injected embryos, and n = 28 for embryos co-injected with N-cadherin MO and constitutively active Src.
(G) Initial frame (0 hr) and last frame (18 hr) from movies of explanted NC cell clusters expressing nuclear RFP, after the indicated treatments. Dispersion analysis based on Delaunay triangles color coded for area size. Scale bar, 100 μm.
(H) Normalized mean triangle areas over time. n = 24 explants for all conditions.
(I) Frames from movies of colliding pairs of NC cells expressing membrane GFP (green) and nuclear RFP (red), after the indicated treatments. Yellow arrows, direction of migration; red arrowhead, cell collision. Scale bar, 10 μm.
(J) CIL within 30 min of colliding, n = 3 repeated experiments for PF-573228 and SU6656. Number of collisions analyzed: control = 250, PF-573228 = 127, SU6656 = 74.
(K) Duration of contact; three repeated experiments for PF-573228 and SU6656. Number of collisions analyzed: control = 250, PF-573228 = 127, SU6656 = 74.
(L) In vivo time-lapse imaging of Sox10-GFP zebrafish embryos showing CIL between NC cells. Yellow arrows, direction of migration; red arrowhead, cell collision; n = 35 embryos.
(M) CIL in vivo within 30 min of colliding, n = 5 repeated experiments for control and DN F FAK. Number of collisions analyzed: control = 150, DN FAK = 117.
(N) Duration of contact. Five repeated experiments for control and DN FAK. Number of collisions analyzed: control = 150, DN FAK = 117.
Line graphs and bar graphs show mean ± SEM. ∗∗∗p ≤ 0.001, ∗∗p ≤ 0.01, ∗p ≤ 0.05. All ANOVA test. See also Figures S4 and S5.