Hybridization of eggs and sperm from closely related species can give rise to genetic diversity, or can lead to embryo inviability due to incompatibility. Although central to evolution, the cellular and molecular mechanisms underlying postzygotic barriers that drive reproductive isolation and speciation remain largely unknown1,2. Species of the African Clawed frog Xenopus provide an ideal system to study hybridization and genome evolution. Xenopus laevis is an allotetraploid with 36 chromosomes that arose through interspecific hybridization of diploid progenitors, whereas Xenopus tropicalis is a diploid with 20 chromosomes that diverged from a common ancestor ~48 million years ago3. Differences in genome size between the two species are accompanied by organism size differences, and size scaling of the egg and subcellular structures such as nuclei and spindles formed in egg extracts4. Nevertheless, early development transcriptional programs, gene expression patterns, and protein sequences are generally conserved5,6. Interestingly, whereas the hybrid produced when X. laevis eggs are fertilized by X. tropicalis sperm (le×ts) is viable, the reverse hybrid (te×ls) dies prior to gastrulation7,8 (Fig. 1a). Here, we applied cell biological tools and high-throughput methods to study the mechanisms underlying hybrid inviability. We reveal that two specific X. laevis chromosomes are incompatible with the X. tropicalis cytoplasm and are mis-segregated during mitosis, leading to unbalanced gene expression at the maternal to zygotic transition, followed by cell-autonomous catastrophic embryo death.
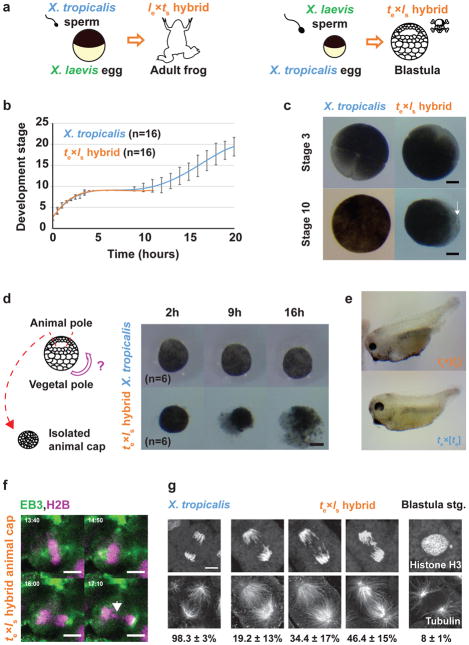
Figure 1. Role of the X. laevis genome in te×ls hybrid embryo death.
a, Schematic of X. laevis and X. tropicalis cross-fertilization outcomes. b, Developmental timing in X. tropicalis and te×ls hybrid embryos. Average is plotted for each time point. Error bars show standard deviation. c, Representative images of X. tropicalis and te×ls hybrid embryos at stages 3 and 10 from experiments in b (n = 16 X. tropicalis and n = 16 te×ls hybrid embryos from 4 independent experiments). The arrow indicates vegetal cells where death initiates. d, Schematic of animal cap assay and images of at 2, 9 and 16 h after isolation. 6 animal caps were imaged and identical results were obtained in 3 different experiments. Scale bars in c and d, 200 μm. e, Images showing haploid phenotype following fertilization of X. tropicalis eggs with UV-irradiated sperm. Identical results were observed in n = 3 experiments. f, Time-lapse images of dividing cell in a te×ls hybrid animal cap (Video 5). The arrow indicates a mis-segregated chromosome. Mis-segregated chromosomes were observed in n = 3 live te×ls hybrid animal caps in 3 experiments. Time is in mm:ss. g, Immunofluorescence images showing chromosome bridges, mis-segregated chromosomes, and micronuclei throughout te×ls hybrid embryos. Scale bars in f and g are 10 μm. Quantification of n = 81 X. tropicalis and n = 78 te×ls hybrid anaphases in n = 17 and 16 embryos, respectively, from 4 datasets obtained from 3 experiments presented as averages ± 1 standard deviation, show a significant difference by Fisher 2 by 3 contingency test (p = 0). Quantification of micronuclei in te×ls hybrid embryos is detailed in Extended Data Figure 1b.
Although te×ls hybrid and X. tropicalis (te×ts) cleavage divisions and rate of development were very similar (Fig. 1b), hybrid embryos died abruptly as late blastulae and never initiated gastrulation. Prior to their death, hybrid embryos took on a deformed mushroom-like shape before lysing from the vegetal pole (Fig. 1c and Video 1). Explants prepared from the opposite pole (animal caps) of mid-blastula te×ls embryos also died within a few hours, indicating that embryo death is cell autonomous and not a result of faulty developmental cues (Fig. 1d and Video 2). In contrast to te×ls hybrids that die as embryos, haploid Xenopus embryos develop to the tadpole stage8,9, suggesting that hybrid death is due to factors brought in by the X. laevis sperm to the X. tropicalis egg during fertilization. Irradiation of X. laevis sperm prior to fertilization, which destroys the DNA10,11, resulted in a haploid phenotype (Fig. 1e and Videos 3, 4), indicating that te×ls embryo death is due to the presence of the X. laevis genome. Cybrid embryos generated by irradiating X. tropicalis eggs, destroying the maternal DNA8 prior to fertilization with X. laevis sperm, died before gastrulation similar to te×ls embryos, indicating that hybrid inviability does not result from a conflict between the paternal and maternal genomes (Extended Data Table 1).
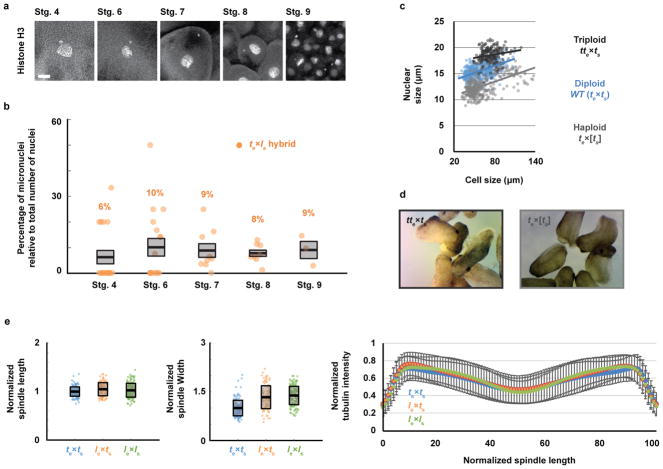
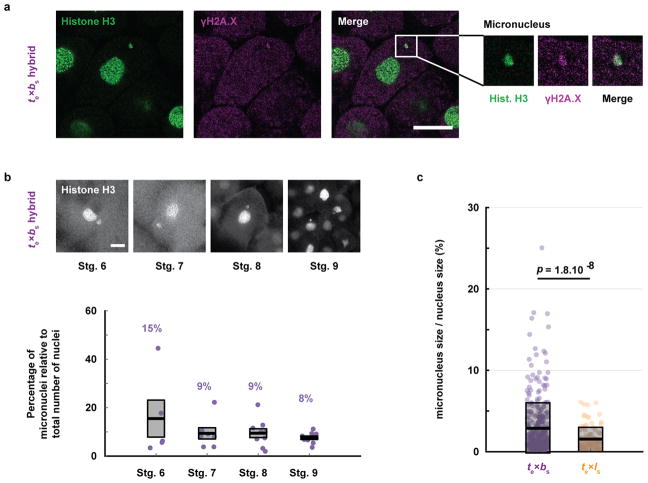
To visualize the dynamics of hybrid cell divisions, we injected mRNAs encoding fluorescent fusion proteins to label embryo chromosomes and mitotic spindles, and observed animal caps at early stage 9, which revealed anaphase defects and chromosome mis-segregation (Fig. 1f and Video 5). Immunofluorescence of whole embryos confirmed the presence of lagging chromosomes and chromosome bridges in cells throughout hybrid blastulae, as well as the formation of micronuclei in interphase, whereas no such defects were observed in X. tropicalis embryos (Fig. 1g) or in the reverse viable hybrid (data not shown). Imaging of te×ls embryos from stage 4 (8 cells) to stage 9 (thousands of cells) revealed micronuclei in 6–10% of the cells throughout hybrid development, but not in the X. tropicalis control (Extended Data Fig. 1a, b), indicating that chromosome mis-segregation in te×ls hybrid embryos is unrelated to changes in gene expression at the onset of zygotic genome activation (ZGA). Since the regular ploidy supported by the X. tropicalis egg is N=20 chromosomes, but the te×ls hybrid zygote must accommodate 28 chromosomes, we tested whether an increase in ploidy was causing chromosome mis-segregation and embryo death by applying a cold shock to X. tropicalis zygotes a few minutes after fertilization to suppress polar body extrusion and increase their ploidy to N=30 chromosomes (Extended Data Fig. 1c). Micronuclei were not observed in cold-shocked embryos, which developed to the tailbud stage similarly to haploid embryos (Extended Data Fig. 1d). Thus, increasing the ploidy of X. tropicalis embryos does not cause chromosome mis-segregation or cell death, indicating a specific role for the X. laevis genome in hybrid inviability.
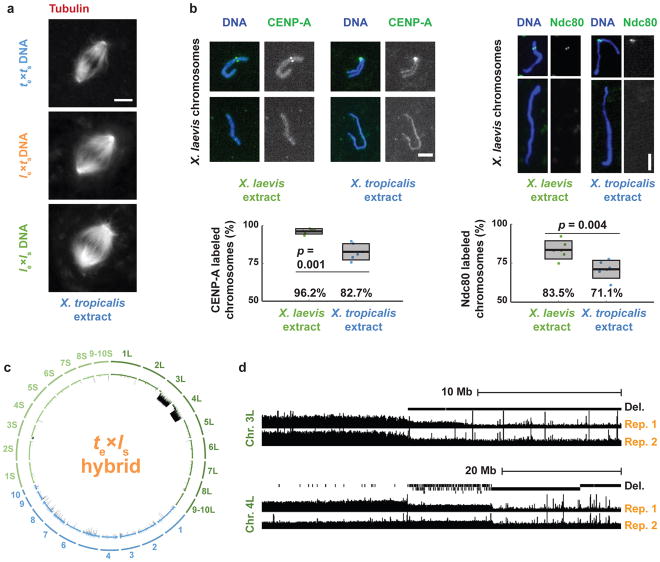
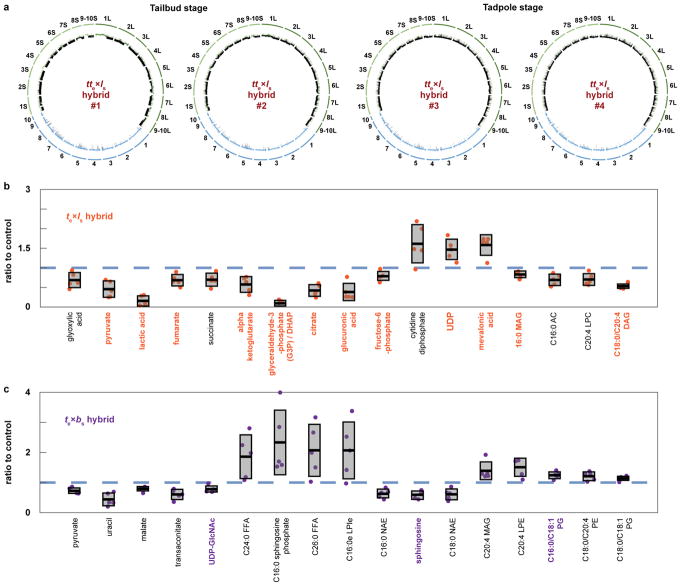
To determine whether assembly and function of the mitotic apparatus was affected, we used the in vitro egg extract system to examine spindle assembly and mitotic chromosome morphology. Metaphase-arrested X. tropicalis egg extract reconstituted spindle formation around nuclei isolated from stage 8 X. tropicalis (N=20), X. laevis (N=36), and viable hybrid embryos (le×ts; N=28) (Fig. 2a). Spindle width scaled slightly with increasing genome size, but microtubule distribution was not affected by either genome size or content (Extended Data Fig. 1e), indicating that the presence of X. laevis DNA did not impair spindle assembly in X. tropicalis cytoplasm. To interrogate chromosome morphology, X. laevis sperm nuclei were cycled through S phase in either X. laevis or X. tropicalis egg extract, induced to arrest in metaphase, and then stained with a DNA dye and antibodies to either CENP-A, the core centromeric histone variant, or Ndc80, an outer kinetochore component essential for linking centromeres to spindle microtubules12. Two fluorescent spots per chromosome were often visible in either extract suggesting that the X. tropicalis extract is capable of replicating the X. laevis genome to generate duplicated sister chromatids. However, we observed 13.5% fewer CENP-A-labeled and 12% fewer Ndc80-labeled chromosomes in X. tropicalis extract compared to X. laevis extract (Fig. 2b), suggesting that approximately two X. laevis chromosomes do not possess centromeres that become competent for kinetochore assembly following a cell cycle in X. tropicalis cytoplasm. Remarkably, whole genome sequencing of embryos at stage 9 prior to cell death revealed the specific loss of 228 Mb of X. laevis sequence from te×ls hybrids (Fig. 2c), 96% of which was missing from just two chromosomes, 3L and 4L. In contrast, no genomic deletions were detected in viable le×ts hybrid embryos (data not shown). Chromosome regions adjacent to breakpoints were heterogeneous in abundance (Fig. 2d), consistent with stochastic chromosome breakage and loss. Notably, major breakpoints localized to a gap in the genome assembly, indicating the presence of repetitive elements. Chromosome loss and partial deletion has been observed in nonviable hybrids in fish13,14 and Drosophila15, but the underlying mechanisms were unclear. Our results suggest that te×ls hybrid incompatibility may be due to divergence of centromeric sequences, which are poorly characterized in Xenopus but known to evolve rapidly16, or to other unidentified repetitive DNA elements that lead to chromosome instability and ultimately prevent kinetochore assembly on chromosomes 3L and 4L.
Figure 2. Compatibility of X. laevis chromosomes with X. tropicalis cytoplasm.
a, Fluorescence images of spindles formed around X. tropicalis, le×ts hybrid, and X. laevis chromosomes in X. tropicalis egg extract. Scale bar, 10 μm. Quantification for n = 147, 103, and 156 spindles quantified for X. tropicalis, le×ts hybrids, and X. laevis embryo nuclei, respectively, from 3 different egg extracts, is presented in Extended Data Figure 1e. b, Fluorescence images of X. laevis chromosomes stained for CENP-A or Ndc80 following replication in X. laevis or X. tropicalis egg extract. CENP-A and Ndc80 labeling was quantified from 6 experiments (3 biological replicates in 2 technical replicates), a total of n = 1792 and n = 1959 chromosomes, respectively in X. laevis extract, and n = 2692 and n = 1930, respectively, in X. tropicalis extract. Scale bars, 5 μm. Box plots show the 6 experiment percentages as individual data points, their average as thick lines, and 1 standard deviation as gray boxes. 95% confidence intervals are 96.2±1.9% in X. laevis extract vs. 82.7±5.7% in X. tropicalis extract for CENP-A and 83.5±6.1% vs. 71.1±6.0% for Ndc80. P-values were determined by two-tailed heteroscedastic t-test. c, Circle plot of whole genome sequencing data for te×ls hybrid embryos aligned and normalized to the genomes of X. tropicalis (blue) and X. laevis (green), with underrepresented genome regions in black. d, Expanded view of chromosome 3L and 4L breakpoints with deleted regions indicated in two biological replicates.
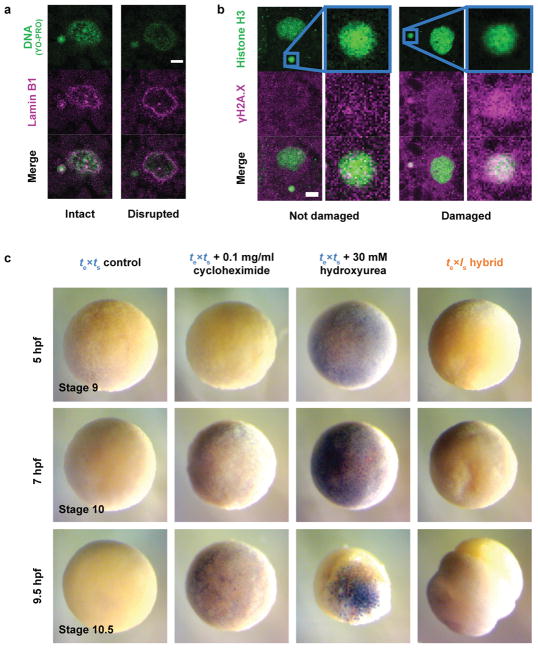
We next investigated the link between chromosome loss and te×ls hybrid embryo death. Micronuclei in cancer cells accumulate DNA damage17–19 and, in Xenopus, DNA damage was shown to trigger apoptosis at the onset of gastrulation20. As in cancer cells, micronuclei in te×ls hybrid embryos often lost envelope integrity and contained damaged DNA (Extended Data Fig. 2a, b). However, te×ls hybrid death did not resemble TUNEL-positive apoptotic death induced by chemical inhibitors of DNA replication or protein synthesis in X. tropicalis embryos (Extended Data Fig. 2c; Videos 6, 7; Extended Data Table 2). We hypothesized that chromosome loss could lead to cell death by affecting gene expression at ZGA. To assess the effects of blocking gene expression globally, we treated X. tropicalis embryos with the transcription initiation inhibitor triptolide and observed a phenotype reminiscent of the timing and manner of the catastrophic te×ls hybrid embryo death, although lysis did not initiate from the vegetal side (Videos 8, 9; Extended Data Table 2). To test whether altering gene dosage could rescue hybrid viability, we applied cold shock to the hybrid zygote to suppress polar body extrusion and introduce a second copy of the X. tropicalis genome. Although extremely inefficient, a total of 9 triploid hybrid tte×ls embryos were obtained in 4 separate experiments and survived to tailbud/tadpole stages (Fig. 3a). Rescued embryos possessed significantly higher DNA content than diploid X. tropicalis embryos at stage 21 (Fig. 3b), but whole genome sequencing revealed that X. laevis DNA was eliminated by the tadpole stage (Extended Data Fig. 3a, Extended Data Table 3, Supplementary Table 1). Our results link te×ls hybrid inviability with altered gene expression that can be rescued with a second copy of the X. tropicalis genome, and indicate that te×ls hybrid embryo inviability is caused by defects at the onset of ZGA, and not by DNA damage and apoptosis.
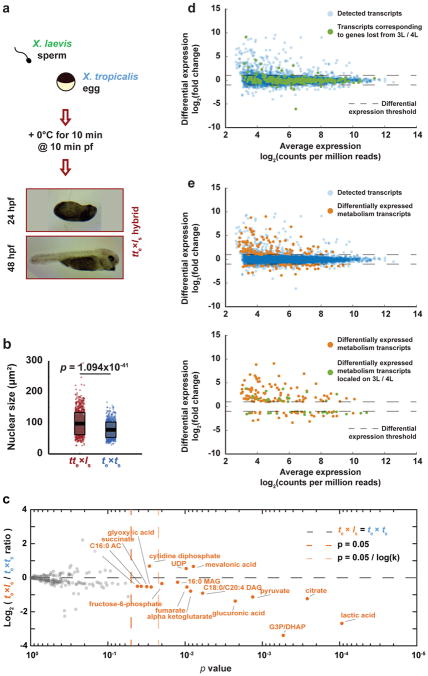
Figure 3. Gene expression and metabolic changes preceding te×ls hybrid embryo death.
a, Schematic of polar body suppression experiment and images of tte×ls rescued embryos 24 h and 48 h post-fertilization (hpf). A total of 9 tte×ls embryos were obtained in 4 different experiments. b, Box plot of nuclear sizes (n = 988 nuclei from 3 tte×ls embryos and n = 777 from 3 X. tropicalis embryos at stage 21) showing the average area as thick lines and 1 standard deviation as gray boxes. 95% confidence intervals are 98.1±2.2 μm2 for tte×ls and 78.0±1.7 μm2 for X. tropicalis embryos. P-values were determined by two-tailed heteroscedastic t-test. c, Levels of 179 metabolites in X. tropicalis and te×ls hybrid embryos 7 hpf. Levels were obtained from 5 samples from 3 independent fertilizations, each averaged and plotted as Log2 of the ratio with the control (see Methods). P-values were calculated using a two-tailed homoscedastic t-test. The average and 1 standard deviation for the differentially represented metabolites are shown, and 95% confidence intervals given in Extended Data Figure 3b. d, Differential gene expression between te×ls and te×ts (see Methods). All detected transcripts (n = 8379) are plotted in blue. Transcripts corresponding to genes lost from chromosomes 3L and 4L (n = 270) are plotted in green. e, Differential expression of metabolism genes between te×ls and te×ts (see Methods). Differentially expressed metabolism transcripts (n = 165) are plotted in orange, all detected transcripts (n = 8379) in blue (top), and differentially expressed metabolism transcripts located on chromosomes 3L and 4L (n = 35) in green (bottom).
Since metabolite pools are known to become crucial before gastrulation21, we subjected te×ls hybrid embryos to metabolic profiling at 7 hours post-fertilization (hpf), just prior to the characteristic deformation preceding lysis. Levels of 17 out of 179 metabolites detected were significantly altered (Fig. 3c). Reduction in lactic acid, the final product of fermentation, and tricarboxylic acid cycle intermediates revealed that glycolytic metabolism was impaired in the cytoplasm of te×ls hybrid embryos, which could in turn alter lipid metabolites including neutral lipids such as diacylglycerols (DAGs) and monoacylglycerols (MAGs), as well as fatty acid oxidation metabolites such as acyl carnitines (ACs) (Extended Data Fig. 3b). While inhibition of mitochondrial ATP synthase led to cell cycle arrest at stage 9 (Video 10), perturbing the early steps of glycolysis in te×ts embryos induced cell death and lysis (Video 11; Extended Data Table 2). In particular, inhibition of glycogen phosphorylase to block the release of glucose from glycogen led to cell death at stage 9, initiating from the vegetal side of the embryo (Video 12). These results are consistent with glycolytic defects as a primary cause of te×ls hybrid embryo death. However, other defects that contribute to hybrid incompatibility could be masked by the abrupt cell lysis, such as conflicts between the paternal genome and maternal mitochondria22,23.
To evaluate the link between the metabolic defects and specific chromosome loss, we used a statistical analysis24 to classify the list of 1803 genes mapped to the regions lost from chromosomes 3L and 4L in te×ls. We found that metabolic processes, particularly in glycolysis, were significantly over-represented (Extended Data Table 4). Transcriptome profiling of te×ls hybrid embryos at 7 hpf (Supplementary Table 2) revealed that although a large fraction of genes lost from chromosomes 3L and 4L were not differentially expressed compared to wild type embryos (>92%; Fig. 3d), 27.1% of the differentially expressed genes related to metabolism (Fig. 3e; top), including PDK1 (pyruvate dehydrogenase kinase). Moreover, 36.7% of the significantly under-expressed metabolism genes are found on chromosomes 3L and 4L (Fig. 3e; bottom), including GFPT1 (fructose-6-phosphate aminotransferase) and HPDL (4-hydroxyphenylpyruvate dioxygenase).
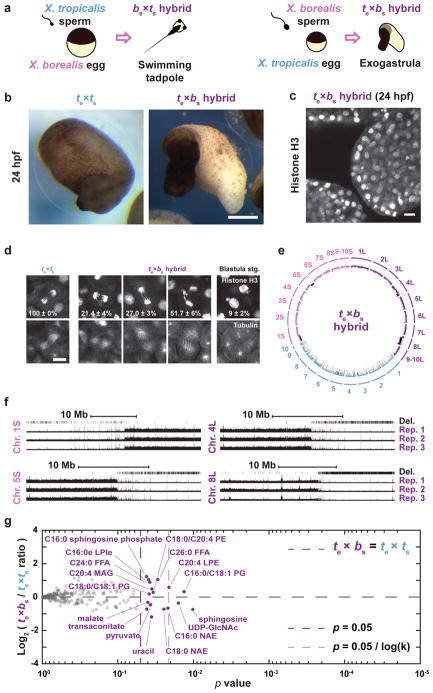
To further characterize the specificity and mechanism underlying te×ls hybrid incompatibility, we compared the outcome of cross-fertilizations between X. tropicalis and another allotetraploid Xenopus species, X. borealis25. Analogous to hybridization between X. laevis and X. tropicalis, we observed that X. borealis eggs fertilized with X. tropicalis sperm (be×ts) were viable while the reverse hybrid (te×bs) was not (Fig. 4a). However, the te×bs embryos did not lyse, but exogastrulated and survived for hours with intact cells (Fig. 4b–c; Video 13). Similar to te×ls, te×bs embryos displayed chromosome loss through anaphase defects and formation of micronuclei (Fig. 4d, Extended Data Fig. 4a–c). Strikingly, whole te×bs hybrid genome sequencing revealed that, although the loss was specific for the paternal genome as in the te×ls hybrid, specific regions of four different X. borealis chromosomes were affected (Fig. 4e–f, Extended Data Table 3, Supplementary Table 1). Furthermore, metabolomics of te×bs embryos revealed a distinct profile with less severe alterations than observed for te×ls (Fig. 4g).
Figure 4. Chromosomal loss in exogastrulating te×bs hybrid embryos.
a, Schematic of X. borealis and X. tropicalis cross-fertilization outcomes. b, Representative images of te×ts vs. te×bs embryos at 24 hpf. This result was reproduced in 4 separate experiments. Scale bar, 200 μm. c, Immunofluorescence image of te×bs hybrid embryo at 24 hpf showing nuclei and micronuclei. Similar defects at this stage were observed in 6 different embryos. d, Immunofluorescence images showing chromosome bridges, mis-segregating chromosomes, and micronuclei throughout te×ls hybrid embryos. Scale bars, 20 μm. Quantification of n = 33 X. tropicalis and 63 te×bs hybrid anaphases in n = 6 and 12 embryos, respectively, show a significant difference by Fisher 2 by 3 contingency test (p = 0). Quantification of micronuclei in te×bs hybrid embryos is detailed in Extended Data Figure 4b. e, Circle plot of whole genome sequencing data for te×bs hybrid embryos aligned and normalized to the genomes of X. tropicalis (blue) and X. borealis (purple). Underrepresented genome regions (black) represent 9.674% of chromosome 4L, 74.66% of 8L, 4.71% of 1S, and 14.4% of 5S. f, Expanded view of chromosome 1S, 5L, 4L and 8L breakpoints with deleted regions indicated in three biological replicates. g, Levels of 241 metabolites in X. tropicalis and te×bs hybrid embryos 7 hpf (see Methods). Levels were obtained from 5 samples from 3 independent fertilizations, each, averaged and plotted as Log2 of the ratio with the control (see Methods). P-values were calculated using a two-tailed homoscedastic t-test. The average and 1 standard deviation for the differentially represented metabolites are shown, and 95% confidence intervals given in Extended Data Figure 3c. Note that few metabolites are altered significantly and are distinct from those altered in te×ls hybrids (see Extended Data Figure 3b–c).
Altogether, our results indicate that hybrid instability in Xenopus results primarily from post-zygotic conflicts between the maternal cytoplasm and the paternal genome that lead to loss of specific genomic regions and downstream gene dosage defects. These findings highlight the role of genome evolution and transmission in defining hybrid fates and speciation.
METHODS
Chemicals
Unless otherwise stated, all chemicals were purchased from Sigma-Aldrich, St. Louis, MO.
Frogs
All animal experimentation is this study was performed according to our Animal Use Protocol approved by the UC Berkeley Animal Care and Use Committee. Mature X. laevis, X. tropicalis, and X. borealis frogs were obtained from NASCO, WI, or the National Xenopus Resource (NXR, Woods Hole, MA). Female X. laevis (1 to 4 years old), X. tropicalis (6 months to 4 years old), and X. borealis (from 2 to 3 years old) frogs were ovulated with no harm to the animals with 6-, 3-, and 4-month rest intervals, respectively. To obtain testes, males (same age ranges) were euthanized by over-anaesthesia through immersion in ddH2O containing 0.15% MS222 (Tricaine) neutralized with 5 mM sodium bicarbonate prior to dissection, and then frozen at −20°C.
Experimental design
Sample sizes were not pre-specified and were sufficient to generate statistically significant differences. All attempts at replication where successful. All experiments were performed independently at least 3 times (biological replicates). Xenopus frogs were selected randomly from our colony for ovulation and fertilization experiments. Investigators were not blinded.
In vitro fertilization and cross-fertilization
X. laevis males were injected with 500 U of human chorionic gonadotropin hormone (hCG) 12–24 h before dissection and testes were stored at 4°C in 1X MR (100 mM NaCl, 1.8 mM KCl, 2 mM CaCl2, 1 mM MgCl2 and 5 mM HEPES-NaOH pH 7.6) for 1–2 weeks. X. tropicalis and X. borealis males were injected with 250 U and 300 U, respectively, of hCG 12–24 h before dissection and testes were collected in Leibovitz L-15 Medium (Gibco – Thermo Fisher Scientific, Waltham, MA) supplemented with 10% Fetal Bovine Serum (FBS; Gibco) for immediate use.
For X. tropicalis egg-based embryos, X. tropicalis females were primed with 25 U of hCG 12–24 h before use and boosted with 250 U of hCG on the day of the experiment. As soon as the first eggs were laid (~3 h after boosting), the X. tropicalis male was euthanized and dissected. Two X. tropicalis or X. borealis testes, or 1/3 of a X. laevis testis were each added to 1 mL of L-15 10% FBS. X. tropicalis females were squeezed gently to deposit eggs onto petri dishes coated with 1.5% agarose in 1/10X MMR (1X MMR: 100 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgSO4 and 5 mM HEPES-NaOH pH 7.6, 0.1 mM EDTA). Testes were homogenized using scissors and a pestle in L-15 10% FBS. Any liquid in the petri dishes was removed and the eggs were fertilized with 500 μL of sperm solution per dish. Eggs were swirled in the solution to separate them and incubated for 4 min with the dish slanted. Dishes were then flooded with ddH2O, swirled and incubated for 5–10 min. Buffer was exchanged for 1/10X MMR, the eggs incubated for 10 min and jelly coats removed with a 3% cysteine solution (in ddH2O-NaOH, pH 7.8). After extensive washing with 1/10X MMR (>4X), embryos were incubated at 23°C. At stage 2–3, fertilized embryos were sorted and placed in fresh 1/10X MMR within new petri dishes coated with 1.5% agarose in 1/10X MMR.
For X. laevis egg-based embryos, X. laevis females were primed with 100 U of pregnant mare serum gonadotropin (PMSG, National Hormone and Peptide Program, Torrance, CA) at least 48 h before use and boosted with 500 U of hCG 14 hours before the experiment. X. laevis females were squeezed gently to deposit eggs onto petri dishes coated with 1.5% agarose in 1/10X MMR. Two X. tropicalis testes collected in L-15 10% FBS or 1/3 of a X. laevis testes were each added to 1 mL of ddH2O and homogenized using scissors and a pestle. Any liquid in the petri dishes was removed and the eggs were fertilized with 500 μL of sperm solution per dish. Eggs were swirled in the solution to individualize eggs as much as possible and incubated for 10 min. Dishes were flooded with 1/10X MMR, swirled and incubated for 10–20 min. Jelly coats were removed with a 2% cysteine solution (in ddH2O-NaOH, pH 7.8). After extensive washing (>4X) with 1/10X MMR, embryos were incubated at 23°C. At stage 2–3, fertilized embryos were sorted and placed in fresh 1/10X MMR in new petri dishes coated with 1.5% agarose in 1/10X MMR.
For X. borealis egg-based embryos, X. borealis females were primed with 60 U of PMSG at least 48 hours before use and boosted with 300 U of hCG 14 h before the experiment. Frogs were kept at 16°C in 1/2X MMR. Eggs were picked from the tub and deposited onto petri dishes coated with 1.5% agarose in 1/10X MMR. Two X. tropicalis or X. borealis testes were collected and homogenized using scissors and a pestle in L-15 10% FBS. Any liquid in the petri dishes was removed and the eggs were fertilized with 500 μL of sperm solution per dish. Eggs were swirled in the solution to individualize eggs as much as possible and incubated for 10 min. Dishes were flooded with 1/10X MMR, swirled and incubated for 10–20 min. Jelly coats were then removed with a 3% cysteine solution (in ddH2O-NaOH, pH 7.8). After extensive washing (>4X) with 1/10X MMR, embryos were incubated at 23°C. At stage 2–3, fertilized embryos were sorted and placed in fresh 1/10X MMR in new petri dishes coated with 1.5% agarose in 1/10X MMR.
All embryos were staged according to Nieuwkoop and Faber26.
Embryo chemical treatments and video imaging
Chemical treatments were performed in petri dishes coated with exactly 5 mL of 1.5% agarose in 1/10X MMR covered with 10 mL 1/10X MMR for either regular incubations or video imaging for consistency. Concentrations were calculated relative to the covering volume of 1/10X MMR, no dilution within the volume in the agarose was assumed. Cycloheximide was added at a concentration of 0.1 mg/mL at stage 6.5 from 8 mg/mL stock in DMSO. Hydroxyurea (Thermo Fisher Scientific, Waltham, MA) was added at a concentration of 30 mM at stage 3 from 600 mM stock in ddH2O. Triptolide was added at a concentration of 25 μM at stage 2 from 25 mM stock in DMSO. Oligomycin was added at a concentration of 40 μM at stage 2 from 40 mM stock in DMSO. AP-III-a4 was added at a concentration of 30 μM at stage 2 from 1 mM stock in DMSO. Iodoacetic acid was added at a concentration of 50 mM at stage 2 from 1 M stock in ddH2O. CP-91,149 was added at a concentration of 270 μM at stage 2 from 30 mM stock in DMSO. Corresponding volumes of DMSO or ddH2O were added to controls.
Imaging dishes were prepared using a homemade PDMS mold designed to print a pattern of 0.9 mm large wells in agarose that allowed us to image 6 X. tropicalis embryos simultaneously within the 3X4 mm camera field of view for each condition. Embryos were imaged from stage 2–3. Treatment and control videos were taken simultaneously using two AmScope MD200 USB cameras (AmScope, Irvine, CA) each mounted on an AmScope SE305R stereoscope. Time lapse movies were acquired at a frequency of 1 frame every 10 s for 20 h and saved as Motion JPEG using a MATLAB (The MathWorks, Inc., Natick, MA) script. Movie post-processing (cropping, concatenation, resizing, addition of scale bar) was done using MATLAB and Fiji27. All MATLAB scripts written for this study are available upon request. Two of the scripts used here were obtained through the MATLAB Central File Exchange: “videoMultiCrop” and “concatVideo2D” by Nikolay S.
Embryo ploidy manipulations
To generate X. tropicalis haploid embryos (te×[ts] and te×[ls]), fertilizations were conducted as detailed above with slight modifications to accommodate for sperm UV-irradiation. Two X. tropicalis testes or 1/3 of a X. laevis testis were each added to 1.1 mL of L-15 10% FBS. Testes were homogenized using scissors and a pestle and the solutions spun briefly using a benchtop centrifuge to pellet the tissue. 1 mL of supernatant was transferred into a glass petri dish and irradiated within a Stratalinker UV-Crosslinker (Stratagene, San Diego, CA) with 50,000 microjoules for X. tropicalis sperm or 2 times 30,000 microjoules for X. laevis sperm, swirling the solution in between the two irradiations. X. tropicalis eggs freshly squeezed onto petri dishes coated with 1.5% agarose in 1/10X MMR were then fertilized with 500 μL of irradiated sperm solution per dish and processed as described above.
To generate [te]×ls cybrid embryos and the haploid [te]×ts controls, fertilizations were conducted as detailed above with slight modifications to accommodate for the UV-irradiation of the eggs. Two X. tropicalis testes or 2/3 of a X. laevis testis were each added to 1.1 mL of L-15 10% FBS. X. tropicalis females were squeezed gently to deposit eggs onto petri dishes coated with 1.5% agarose in 1/10X MMR. Excess liquid was removed, eggs were swirled with a pestle to form a monolayer of properly oriented eggs and immediately irradiated within a Stratalinker UV-Crosslinker (Stratagene, San Diego, CA) 2 times with 40,000 microjoules. Testes were homogenized using scissors and a pestle during the irradiation of the eggs. As soon as irradiated, the eggs were fertilized with 500 μL of sperm solution per dish and processed as described above.
To prevent polar body formation in either tte×ts or tte×ls experiments, fertilizations were conducted as detailed above with slight modifications to accommodate cold treatment. Fertilizations were performed within dishes coated with only 1–1.5 mL, instead of 5 mL, of 1.5% agarose in 1/10X MMR to accelerate cooling. Following the 4-min incubation with sperm, dishes were flooded with ddH2O, swirled and incubated for exactly 5 min. Buffer was then exchanged for ice-cold 1/10X MMR, the dishes transferred into a pipette tip box lid placed in a slushy ice bucket, and the eggs were incubated for 10 min. The dishes were then removed from the bucket and the cold buffer was exchanged for RT 1/10X MMR. After 20 min, the jelly coat was removed with a 3% cysteine solution (in ddH2O-NaOH, pH 7.8) and the embryos processed as described above.
Animal cap assay
At stage 8, embryos were placed in Danilchik’s for Amy Medium (DFA medium; 53 mM NaCl, 5 mM Na2CO3, 4.5 mM Potassium gluconate, 32 mM Sodium gluconate, 1 mM CaCl2, 1 mM MgSO4, pH 8.3, 1 g/L BSA and 0.8% Antibiotic Antimycotic Solution) for surgery. Using Dumostar-Biology 55 forceps (Dumont, Montignez, Switzerland), the vitelline membrane was removed and the animal cap was isolated from the embryo. The caps were finally transferred to a new dish or a chamber containing fresh DFA medium for imaging.
mRNA, embryo microinjection, and animal cap confocal microscopy
Plasmids for expression of EB3-GFP and histone H2B-RFP mRNAs were obtained at the 2013 Advanced Imaging in Xenopus Workshop from the Wallingford lab (UT Austin, USA). The mRNAs were synthetized using mMessage mMachine SP6 Transcription Kit (Ambion – Thermo Fisher Scientific, Waltham, MA) following supplier protocol. The mRNAs were purified using Phenol-Chloroform extraction, resuspended in ddH2O, aliquoted and stored at −80°C.
At stage 2, te×ls hybrid embryos were transferred to 1/9X MMR 3% Ficoll. A solution containing 50 pg/nL of H2B-RFP mRNA and 100 pg/nL of EB3-GFP mRNA, concentrations which allowed us to image fluorescent signal as early as stage 9, was loaded into a needle pulled from 1 mm glass capillary tube (TW100F-4, World Precision Instruments, Inc., Sarasota, FL) using a P-87 Micropipette Puller (Sutter Instrument, Navato, CA). Embryos were placed in a mesh-bottomed dish and microinjected in both blastomeres with 1 nL of the mRNA solution using a Picospritzer III microinjection system (Parker, Hollis, NH) equipped with a MM-3 micromanipulator (Narishige, Amityville, NY). Injected embryos were transferred to a new dish and incubated at 23°C in 1/9X MMR 3% Ficoll until stage 8 when they were processed for animal cap isolation as described above. Caps were placed in a chamber filled with DFA medium made using 1×1 cm Gene Frames (Thermo Fisher Scientific, Waltham, MA) between a slide and a coverslip (Thermo Fisher Scientific, Waltham, MA) for confocal microscopy.
Embryo whole mount immunofluorescence
At desired stages, embryos were fixed for 1–3 h using either MAD fixative (2 parts of methanol (Thermo Fisher Scientific, Waltham, MA), 2 parts of acetone (Thermo Fisher Scientific, Waltham, MA), 1 part of DMSO) for most antibodies or MEMFA fixative (0.1 M MOPS pH 7.4, 2 mM EGTA, 1 mM MgSO4, 3.7% formaldehyde) for the γH2A.X antibody. After fixation, embryos were dehydrated in methanol and stored at −20°C. Embryos were then processed as previously described28 with some modifications. Following gradual rehydration in 0.5X SSC (1X SSC: 150 mM NaCl, 15 mM Na citrate, pH 7.0), embryos were bleached with 1–2% H2O2 (Thermo Fisher Scientific, Waltham, MA) in 0.5X SSC containing 5% formamide for 2–3 h under light, then washed in PBT, a PBS solution containing 0.1% Triton X-100 (Thermo Fisher Scientific, Waltham, MA) and 2 mg/mL bovine serum albumin (BSA). Embryos were blocked in PBT supplemented with 10% goat serum (Gibco – Thermo Fisher Scientific, Waltham, MA) and 5% DMSO for 1–3 h and incubated overnight at 4°C in PBT supplemented with 10% goat serum and the primary antibodies. We used different combinations of the following antibodies: 1:500 mouse anti-beta tubulin (E7; Developmental Studies Hybridoma Bank, Iowa City, IA), 1:500 rabbit anti-histone H3 (ab1791; Abcam, Cambridge, MA), 1:500 rabbit anti-lamin B1 (ab16048; Abcam, Cambridge, MA), 1:500 mouse anti-phospho-histone H2A.X (05-636; EMD Millipore, Merck KGaA, Darmstadt, Germany). Embryos were then washed 4 X 2 h in PBT and incubated overnight in PBT supplemented with 1:500 goat anti-mouse or goat anti-rabbit secondary andibodies coupled either to Alexa Fluor 488 or 568 (Invitrogen – Thermo Fisher Scientific, Waltham, MA) and with 1:200 YO-PRO iodide (Thermo Fisher Scientific, Waltham, MA) if the use of anti-histone H3 antibody as primary was not possible. Embryos were then washed 4 X 2 h in PBT and gradually dehydrated in methanol. Embryos were finally cleared in Murray’s clearing medium (2 parts of Benzyl Benzoate, 1 part of Benzyl Alcohol). Embryos were placed either in a chamber made using a flat nylon washer (Grainger, Lake Forest, IL) attached with nail polish (Sally Hansen, New York, NY) to a slide and covered by a coverslip or a chamber made of silicon grease (Beckman coulter, Brea, CA) between slide and coverslip, and filled with Murray’s clearing medium for confocal microscopy.
Confocal microscopy, micronuclei and nuclear size quantification
Confocal microscopy was performed on a Zeiss LSM 780 NLO AxioExaminer using the Zeiss Zen Software. For animal cap live imaging, histone H2B-RFP and EB3-GFP signals were imaged on a single plane with a frame size of 1024×1024 px every 5 s using a Plan-Apochromat 40x/1.4 Oil objective and laser power of 22%. For imaging of histone H3, embryos were imaged using a Plan-Apochromat 20x/1.0 Water objective and laser power of 12%, on multiple 1024×1024 px plans spaced of 0.68 μm in Z. For characterization of the micronuclei (lamin B1 and γH2A-X), embryos were imaged using a Plan-Apochromat 63x/1.40 Oil objective and laser power of 12%, on multiple plans spaced 0.38 μm in Z. Images are mean averages of 2 scans with a depth of 16 Bits. Pinhole size was always chosen to correspond to 1 airy unit.
Micronuclei were quantified at stages 4, 6, 7, 8 and 9 as the number of observed micronuclei in the dataset divided by the number of nuclei in the dataset. The number of micronuclei at all stages and of nuclei at stage 4 and 6 were counted manually in Fiji. The number of nuclei at stages 7, 8 and 9 was determined automatically through histone H3 fluorescence signal segmentation using Imaris (Bitplane, Zurich, Switzerland). Nuclear area in tte×ts, X. tropicalis and te×[ts] was measured in Fiji using the ellipse tool. From this, we calculated the diameter of a circle of the same area, a value that we could directly compare the cell size determined through the measurement of the cell diameter at the nucleus central plan.
Embryo nuclei purification
Embryo nuclei were prepared as previously described29 from X. tropicalis, le×ts hybrid, and X. laevis embryos. Briefly, embryos were arrested at stage 8 in late interphase using 150 μg/mL cycloheximide in 1/10X MMR for 60 min. Then they were washed several times in ELB (250 mM sucrose, 50 mM KCl, 2.5 mM MgCl2, and 10 mM HEPES pH 7.8) supplemented with LPC (10 μg/mL each leupeptin, pepstatin, chymostatin), cytochalasin D (100 μg/mL), and cycloheximide (100 μg/mL), packed in a tabletop centrifuge at 200 g for 1 min, crushed with a pestle, and centrifuged at 10,000 g for 10 min at 16°C. The cytoplasmic extract containing endogenous embryonic nuclei was collected, supplemented with 8% glycerol, aliquoted, frozen in liquid nitrogen and stored at −80°C.
Xenopus egg extracts and related methods
X. laevis30 and X. tropicalis4 metaphase arrested egg extracts were prepared and spindle reactions conducted as previously described.
To reconstitute spindle assembly, stage 8 embryo nuclei were used as a source of DNA. Aliquots were thawed, resuspended in 1 mL of ELB, and spun at 1600 g for 5 min at RT. Pelleted nuclei were resuspended in 25 μL of fresh X. tropicalis extract and incubated at RT. To examine kinetochore assembly, X. laevis sperm nuclei, prepared as previously described31, were used as a source of DNA in both X. laevis and X. tropicalis egg extracts. Cycled chromosomes were prepared and spun-down30 and then processed for immunofluorescence as previously described32. Briefly, the coverslips were incubated for 1 min in cold methanol, washed with PBS+NP40 and blocked overnight in PBS + 5% BSA at 4°C. The anti-Ndc80 (1:300 dilution, Stukenberg lab, University of Virginia) or the anti-CENP-A (1:500 dilution, Straight lab, Stanford University) rabbit antibodies were added for 1 h. After washing with PBS+NP40, the coverslips were incubated with 1:1000 anti-rabbit antibody coupled to Alexa Fluor 488 (Invitrogen – Thermo Fisher Scientific, Waltham, MA) for 30 min and then with 1:1000 Hoechst for 5 min. The coverslips were finally washed and mounted for imaging. Each presented dataset was obtained from 3 different egg extracts with technical duplicates for each. Spindles and chromosomes were imaged using micromanager software33 with an Olympus BX51 microscope equipped with an ORCA-ER or an ORCA-II camera (Hamamatsu Photonics, Hamamatsu city, Japan), and with an Olympus UPlan FL 40X air objective. Spindle measurements were made using Fiji and the spindle tubulin intensity line scan using an automated Java ImageJ plugin developed by Xiao Zhou (Heald lab, UC Berkeley; https://github.com/XiaoMutt/AiSpindle).
TUNEL assay
Embryos were fixed in MEMFA as described for embryo whole mount immunofluorescence and processed as previously described20 with minor modifications. Briefly, after gradual rehydration, embryos were bleached with 1–2% H2O2 in 0.5X SSC containing 5% formamide for 1–2 h under light. After washes in PBS, embryos were incubated in 1X Terminal Deoxynucleotidyl Transferase (TdT) Buffer for 1 h and then overnight in TdT Buffer supplemented with 150 U/ml of TdT enzyme (Invitrogen – Thermo Fisher Scientific, Waltham, MA) and 1 pmol/μL Digoxigenin-11-dUTP (Roche, Basel, Switzerland). After washes in 1 mM EDTA/PBS at 65°C, in PBS and then in MAB (100 mM maleic acid, 150 mM NaCl, pH 7.5), embryos were blocked for 1 h in 2% Blocking Reagent (Roche, Basel, Switzerland) in MAB and then incubated overnight at 4°C in 2% Blocking Reagent in MAB supplemented with 1:3000 anti-Digoxigenin AP antibody (Roche, Basel, Switzerland). After washes in MAB and in AP Buffer (100 mM Tris, pH 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween 20, 2 mM Levamisol), embryos were stained with NBT/BCIP (nitro-blue tetrazolium/5-Bromo-4-chloro-3-indolyl phosphate; Promega, Sunnyvale, USA) diluted in AP buffer. Reactions were stopped in MAB and embryos fixed overnight in Bouin’s solution. After washes in 70% buffered ethanol and in methanol, embryos were imaged in methanol with the ToupView software (ToupTek, Zhejiang, China) using an AmScope MD200 USB camera mounted on M5 stereoscope (Wild Heerbrugg, Gais, Switzerland).
Nucleic acid isolation, library construction, and sequencing
For genomic DNA, embryos at desired stages were incubated overnight in lysis buffer (50 mM Tris-HCl, 5 mM EDTA, 100 mM NaCl, 0.5% SDS) containing 250 μg/mL Proteinase K (Roche, Basel, Switzerland). DNA was isolated using Phenol-Chloroform extraction and ethanol precipitation. To isolate RNAs, embryos at desired stages were homogenize mechanically in TRIzol® (Thermo Fisher Scientific, Waltham, MA) using up to a 30-gauge needle and processed according to supplier instructions. After resuspension in nuclease-free H2O, RNAs were cleaned up using RNeasy kit (Qiagen Inc.) with on-column DNA digestion, following supplier protocol.
Libraries were constructed at the Functional Genomics Lab (FGL), a QB3-Berkeley Core Research Facility at UC Berkeley. For genomic DNA, an S220 Focused-Ultrasonicator (Covaris®) was used to fragment DNA. The fragmented DNA was cleaned and concentrated with the MinElute® PCR Purification kit (Qiagen Inc.). The Library preparation was done on an Apollo 324™ with PrepX™ ILM 32i DNA Library Kits (WaferGen Biosystems, Fremont, CA), and 7 cycles of PCR amplification was used for library fragment enrichment. For RNAs, mRNA enrichment was performed on total RNA using polyA selection with the Invitrogen Dynabeads mRNA Direct kit. The library preparation was done on the Apollo324™ with PrepX™ RNAseq Library Prep Kits (WaferGen Biosystems, Fremont, CA), and 13 cycles of PCR amplification was used for index addition and library fragment enrichment.
Sequencing was performed by the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley. All samples have been run as 100 paired-end HiSeq4000 lanes, pooled equimolar after quantification using KAPA Illumina Library quantification qPCR reagents on the BioRad CFX connect. Demultiplexing was performed to allow a single mismatch with Illumina’s bcl2fastq version 2.17 software.
Genomic DNA sequencing analysis and deletion detection in hybrids
DNA sequencing reads were mapped to a X. laevis - X. tropicalis hybrid genome (Xenla9.1 and Xentro9.0) or a X. borealis - X. tropicalis hybrid genome (Xbo_04Apr2017 (Mudd and Rokhsar, unpublished) and Xentro9.0) using bwa mem (version 0.7.10-r789) with default settings. Duplicate reads were marked using bamUtil v1.0.2.
Deletions in te×ls and le×ts hybrids were called by comparing local DNA sequencing read coverage between hybrid (te×ls or le×ts) and parental (te×ts or le×ls) genomes. The read coverage was determined in 10kb regions, with a 2kb sliding window across the genome. For each 10kb region we calculated the RPKM in te×ls, le×ts, te×ts and le×ls sequencing tracks. The ratio of median RPKM values in retained regions has a non-zero baseline as expected because of a different size of hybrid and parental genomes. The ratio cut-off for deleted regions was set accordingly at 4-fold and 6-fold for X. laevis and X. tropicalis sequences, respectively. Lost regions overlapping for more than 30% of their length with gaps were removed and regions within 10kb of each other were merged. Lost genes were analyzed using the PANTHER database24. Because PANTHER only provided adjusted p-values for fold enrichment based on binomial test, we additionally estimated the 95% confidence interval for each enriched pathways using binom.test() function in R (version 3.2.2).
Deletions in te×bs hybrids were called by identifying reduced genomic DNA signal in te×bs. The RPKM read coverage was determined in 10kb regions, with a 2kb sliding window across the genome. Regions with median log10 RPKM less than -1.25 in the X. tropicalis genome and -1.15 in the X. borealis genome were marked as deleted. Lost regions overlapping for more than 30% of their length with gaps were removed and regions within 10kb of each other were merged.
RNA sequencing analysis
We mapped RNA-seq reads to the combined primary transcripts of X. laevis (JGIv18pV4) and X. tropicalis (JGIv91) using bwa mem (version 0.7.10), and discarded all reads mapped in multiple targets for further analysis. To use human gene annotation, which is more comprehensive than Xenopus, we transferred the expression level of these species to human orthologs. Based on the best BLASTP hit to human longest protein sequences (from EnsEMBL version 80), we merged the read counts of X. laevis and X. tropicalis genes to corresponding human genes, then performed differential expression analysis with EdgeR34. An adjusted P-value criterion of less than 0.05 was applied to determine the significance of differential expression. To estimate the 95% confidence interval of log-scale fold change, we used limma35 (version 3.28.10). Results of both EdgeR and Limma analyses are presented in Supplementary Table 2. For metabolic gene analysis, we used the list of metabolic genes obtained from the PANTHER database24.
Metabolomic profiling
Seven hours post-fertilization, te×ls hybrid, te×bs hybrid, and respective X. tropicalis control embryos were collected from 3 independent fertilizations, always using eggs from the same female between the te×ls hybrid or the te×bs hybrid, and its X. tropicalis control. Five samples of 8 embryos each for nonpolar lipid metabolites and 12 embryos each for polar metabolites were rinsed twice in filtered PBS and frozen in liquid nitrogen. Nonpolar lipid metabolites from the 8 embryos were extracted in 3 mL of 2:1 chloroform:methanol and 1 mL of PBS with inclusion of internal standards C12 monoalkylglycerol ether (MAGE) (10 nmol, Santa Cruz Biotechnology) and pentadecanoic acid (10 nmol). Organic and aqueous layers were separated by centrifugation at 1000 g for 5 min and the organic layer was collected, dried under a stream of nitrogen and dissolved in 120 μl chloroform. Polar metabolites were extracted from the 12 embryos in 180 μL of 40:40:20 (ACN:MeOH:H2O) with inclusion of internal standard D3N15 serine (50 nM, Cambridge Isotope Laboratories, Inc. #DNLM-6863). Samples were disrupted by sonication then centrifuged at 21,000 g for 10 min and the supernatant was collected for analysis. Metabolites were separated by liquid chromatography and MS analysis was performed with an electrospray ionization (ESI) source on an Agilent 6430 QQQ LC-MS/MS (Agilent Technologies). The capillary voltage was set to 3.0 kV, and the fragmentor voltage was set to 100 V. The drying gas temperature was 350°C, the drying gas flow rate was 10 l/min, and the nebulizer pressure was 35 psi. Metabolites were identified by single reaction monitoring (SRM) of the transition from precursor to product ions at associated optimized collision energies and retention times as previously described36. Metabolites were quantified by integrating the area under the curve, then normalized to internal standard values and tissue weight. Metabolite levels are expressed as relative abundances as compared to controls. P-values were calculated using a two-tailed homoscedastic t-test. Significance was analyzed for an α = 0.05 threshold as well as that with a Bonferroni-like correction to account for multiple hypothesis comparison. Strict Bonferroni correction is highly conservative and often results in increased type II errors (failing to acknowledge a real effect) and several alternatives exist. Because we compared around 200 compounds between two types of embryos, each with 5 replicates, we used a penalized Bonferroni correction, and divided the α threshold value by the logarithm of the number of tests (α/log(k)) to decrease the risk for the type II error.
Data availability
All genomic and transcriptomic data generated for this study are available from public databases: stage 9 te×ls hybrid whole genome sequencing data (NCBI SRA: SRP124316; https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP124316) and corresponding stage 9 X. laevis, X. tropicalis and le×ts hybrid controls (NCBI GEO: GSE92382; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE92382), tailbud and tadpole stage tte×ls hybrid whole genome sequencing data (NCBI SRA: SRP124316; https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP124316), stage 9 te×bs hybrid whole genome sequencing data (NCBI SRA: SRP124316; https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP124316) and 7 hpf X. tropicalis and te×ls hybrid transcriptome RNA-seq data (NCBI GEO: GSE106157; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE106157).
Code availability
MATLAB scripts were written to acquire and process embryo live imaging movies. All these scripts are available from the corresponding author upon request. Two MATLAB scripts used here for movie cropping and concatenation were obtained through the MATLAB Central File Exchange: “videoMultiCrop” and “concatVideo2D” by Nikolay S. The automated spindle tubulin intensity line scan Java ImageJ plugin developed by Xiao Zhou (Heald lab, UC Berkeley) is available on GitHub (https://github.com/XiaoMutt/AiSpindle).
Extended Data
Extended Data Figure 1. Occurrence of micronuclei, role of ploidy and spindle architecture.
a, Micronuclei in te×ls hybrid embryos at various developmental stages. Whole mount embryo immunofluorescence was performed in te×ls hybrid embryos using anti-histone H3 antibody at stages 4, 6, 7, 8 and 9 and quantified in b. Scale bar is 10 μm. b, Quantification of micronuclei in te×ls hybrid embryos. The percentage of micronuclei was calculated as the number of micronuclei in the imaged portion of the embryo divided by the total number of nuclei in the same imaged portion. The average percentage for multiple embryos at stage 4 (n = 18 te×ls hybrid embryos (individual dots) with a total of 63 nuclei), stage 6 (n = 17/115), stage 7 (n = 9/322), stage 8 (n = 8/1119) and stage 9 (n = 3/2004) from 3 independent experiments, is shown as thick line. Gray boxes indicate a 1 standard error of the mean. Control X. tropicalis embryos from the same mothers were analyzed but no micronuclei were observed at any stages. c, Nuclear size in X. tropicalis embryos with varying ploidy. Nuclear size relative to cell size (diameters in μm) is plotted for triploid (tte×ts; dark grey, n = 175 nuclei from 6 embryos), diploid (X. tropicalis, te×ts; blue, n = 453/9) and haploid (te×[ts]; light grey, n = 346/16) embryos. Each dot indicates an individual data point and the solid lines indicate a linear fit. d, X. tropicalis embryos with varying ploidy at tailbud stage. Images of triploid (tte×ts; left) and haploid (te×[ts]; right) tailbuds were taken under identical conditions. Similar observations were over 3 independent experiments. e, Size and microtubule distribution in X. tropicalis spindles assembled from different embryo nuclei DNA (n = 147, 103, and 156 spindles quantified for X. tropicalis, le×ts hybrids, and X. laevis embryo nuclei, respectively, from 3 different egg extracts). Spindle length (left) and width (middle) were normalized to the X. tropicalis control, averages are shown as thick black lines and the gray boxes indicate 1 standard deviation. 95% confidence intervals for lengths are 1±0.02 for te×ts, 1.05±0.03 for le×ts, and 1.03±0.02 for le×ls and for widths are 1±0.04, 1.3±0.07, and 1.4±0.04. Line scans of rhodamine-tubulin signal along spindle length were taken (right). Spindle lengths were normalized to 100% and tubulin intensities were normalized within datasets. The average intensities are plotted for the three spindle types, error bars indicate standard deviation and colors are as in Figure 2a.
Extended Data Figure 2. Characterization of micronuclei in te×ls hybrid embryos and link to embryo death.
a, Disrupted micronuclei envelopes in te×ls hybrid embryos. Whole mount embryo immunofluorescence was performed in te×ls hybrid embryos using the YO-PRO DNA dye (top) and anti-Lamin B1 antibody (middle), corresponding channels are shown in green and magenta, respectively. The merged images are shown below. 25 micronuclei within 5 different embryos were analyzed. Intact (left) and disrupted (right) envelopes were observed in all analyzed embryos. Scale bar is 10 μm. b, DNA damage in te×ls hybrid embryo micronuclei. Whole mount embryo immunofluorescence was performed in te×ls hybrid embryos using anti-histone H3 (top) and anti-γH2A.X (middle) antibodies, corresponding channels are shown in green and magenta, respectively. The merge images are shown below. 21 micronuclei within different 6 embryos were analyzed. Micronuclei with undamaged (left; negative γH2A.X signal) and damaged (right; positive γH2A.X signal) DNA were observed in all analyzed embryos. Zoomed images of micronuclei are shown on the right of each image. Scale bar is 10 μm. c, TUNEL assay in apoptotic X. tropicalis and te×ls hybrid embryos. X. tropicalis (left), X. tropicalis treated with cycloheximide (middle left) or hydroxyurea (middle right) as indicated, and te×ls hybrid (right) embryos were prepared for TUNEL assay 5 hpf (equivalent stage 9; top), 7 hpf (equivalent stage 10; middle) and 9.5 hpf (equivalent stage 10.5; bottom). Identical results were obtained over 3 different experiments. Representative images are shown and were taken under identical conditions.
Extended Data Figure 3. Whole genome sequencing of tte×ls rescued embryos and metabolomic profiling of te×ls and te×bs hybrid embryos.
a, The genomes of 4 tte×ls rescued embryos were sequenced, aligned, and normalized to the genomes of X. tropicalis (blue) and X. laevis (green) for which sub-genomes S and L were distinguished (S in light green and L in dark green). Underrepresented regions of the genomes are color-coded in black. The tte×ls embryo genomes 1–2 were prepared from tailbuds, and 3–4 from tadpoles. b, Metabolites differentially represented between te×ls hybrid and X. tropicalis embryos 7 h post fertilization. Among the 179 metabolites detected, 17 were significantly altered in te×ls hybrid embryos (p < 0.05; two-tailed homoscedastic t-test; individual p-values are provided in Figure 3c source data) and are shown as a ratio to the X. tropicalis control (blue dashed line). Levels were obtained from 5 samples from 3 independent fertilizations each. Values for the te×ls hybrid are plotted in orange. The averages are shown as thick lines and the gray boxes correspond to 1 standard deviation. 95% confidence intervals are, from left to right, 0.69±0.24, 0.46±0.26, 0.16±0.16, 0.68±0.18, 0.70±0.21, 0.58±0.25, 0.10±0.09, 0.42±0.19, 0.38±0.27, 0.79±0.15, 1.61±0.61, 1.47±0.33, 1.58±0.33, 0.83±0.11, 0.71±0.18, 0.70±0.19, and 0.53±0.08. Metabolites with p values below the penalized Bonferroni corrected threshold (n = 12) are labeled in orange. c, Metabolites differentially represented between te×bs hybrid and X. tropicalis embryos 7 h post fertilization. Among the 241 metabolites detected, 17 were significantly altered in te×bs hybrid embryos (p < 0.05; two-tailed homoscedastic t-test; individual p-values are provided in Figure 4g source data) and are shown as a ratio to the X. tropicalis control (blue dashed line). Levels were obtained from 5 samples from 3 independent fertilizations, each. Values for the te×bs hybrid are plotted in purple. The averages are shown as thick lines and the gray boxes correspond to 1 standard deviation. 95% confidence intervals are, from left to right, 0.73±0.12, 0.44±0.26, 0.80±0.10, 0.61±0.21, 0.78±0.14, 1.86±0.9, 2.33±1.33, 2.07±1.07, 2.07±1.17, 0.63±0.19, 0.59±0.16 0.61±0.22, 1.39±0.37, 1.51±0.38, 1.24±0.14, 1.21±0.18, and 1.14±0.10. Metabolites with p values below the penalized Bonferroni corrected threshold (n = 3) are labeled in purple.
Extended Data Figure 4. Characterization of micronuclei in te×bs hybrid embryos.
a, DNA damage in te×bs hybrid embryo micronuclei. Whole mount embryo immunofluorescence was performed in te×bs hybrid embryos using anti-histone H3 (left) and anti-γH2A.X (middle) antibodies, corresponding channels are shown in green and magenta, respectively. The merged image is shown on the right. 34 micronuclei within 8 different embryos were analyzed. Micronuclei with damaged DNA were observed in all analyzed embryos. Zoomed images of micronuclei are shown on the right in the same left-to-right order. Scale bar is 20 μm. b, Micronuclei in te×bs hybrid embryos at various developmental stages (top). Whole mount embryo immunofluorescence was performed in te×bs hybrid embryos using anti-histone H3 antibody at stages 6, 7, 8 and 9. Scale bar is 20 μm. Quantification of micronuclei in te×bs hybrid embryos (bottom). The percentage of micronuclei was calculated as the number of micronuclei in the imaged portion of the embryo divided by the total number of nuclei in the same imaged portion. The average percentage for multiple embryos at stage 6 (n = 5 te×bs hybrid embryos (individual dots) with a total of 125 nuclei), stage 7 (n = 7/153), stage 8 (n = 9/731) and stage 9 (n = 10/2691) is shown as a thick line. Gray boxes correspond to 1 standard error of the mean. Control X. tropicalis embryos from the same mothers were analyzed but no micronuclei were observed at any stages. c, Micronuclei size in te×bs and te×ls hybrids. Size is plotted as the ratio between the volumes of the micronucleus and its corresponding nucleus. Each dot represents an individual data point (n = 329 micronuclei from 36 te×bs embryos shown in purple and n = 100 from 17 te×ls embryos shown in orange, from 4 independent experiments). The thick black line indicates the average and the grey box corresponds to 1 standard deviation. 95% confidence intervals are 2.9±0.36% for te×bs and 1.6±0.28% for te×ls embryos. Statistical significance was shown using a two-tailed heteroscedastic t-test.
Extended Data Table 1. Embryonic development in Xenopus haploids and cybrids generated from X. tropicalis irradiated eggs.
n indicates the number of different male-female combinations from which results were compiled. Unfertilized eggs and embryos that showed an abnormal or incomplete first cleavage were excluded from this analysis (†). Fertilization efficiency of irradiated X. tropicalis eggs with X. laevis sperm was very low (~4%) (‡).
| Embryo | Normal Stage 2†(n) | Regular Stage 9 (%) | Died between 9–13 (%) | Exogastrulae | Normal tailbuds | Abnormal tailbuds | Tadpoles | |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Stunted (%) | Normal (%) | |||||||
| [te]×ts | 402 (5) | 402(100) | 6(1) | 131 (33) | 209 (52) | 56 (14) | 191 (48) | 18(4) |
| [te]×ls‡ | 25(7) | 25(100) | 25(100) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) |
Extended Data Table 2. Effects of drug treatments on te×ts embryos.
X. tropicalis embryos were treated with different drugs at different stages. Phenotypes of effects are listed. When unspecified, apoptosis or lysis initiated at random locations in the embryo.
| Drug | Inhibition | Time of addition | Concentration | Phenotype | Product details |
|---|---|---|---|---|---|
| Cycloheximide | Protein synthesis | stage 6.5 | 0.1 mg/ml | Cell cycle arrest at stage 7 followed by apoptosis | C7698 (Sigma-Aldrich) |
| Hydroxyurea | DNA replication | stage 3 | 30 mM | Apoptosis at late stage 8 | AC151680050 (Thermo Fisher Sc.) |
| Triptolide | Transcription | stage 2 | 25 μM | Cell lysis at stage 9 | T3652 (Sigma-Aldrich) |
| Olygomycin | ATP Synthase | stage 2 | 40 μM | Cell cycle arrest at stage 9 | 75351 (Sigma-Aldrich) |
| AP-III-a4 | Enolase (including non-glycolytic functions) | stage 2 | 30 μM | Arrest at stage 7 and followed by cell lysis | 19933 (Cayman Chemical) |
| Iodoacetic acid | Glyceraldehyde-3-P dehydrogenase | stage 2 | 50 mM | Cell lysis at stage 9 | 14386 (Sigma-Aldrich) |
| CP-91,149 | Glycogen phosphorylase | stage 2 | 270 μM | Cell death at stage 9 from the vegetal side | PZ0104 (Sigma-Aldrich) |
Extended Data Table 3. Sub genome distribution of lost vs. retained DNA in te×ls, tte×ls, and te×bs hybrids.
Percentage of lost and remaining DNA for each sub genome is shown for all hybrid genomes sequenced. Sub genomes are color-coded as in Figures 2c and 4e.
| te×ls hybrid | |||||
| Sub genome | Total (bp) | Lost (bp) | Remaining (bp) | Lost (%) | Remaining (%) |
| X laevis L | 1368982762 | 237294229 | 1131688533 | 17.33 | 82.67 |
| X. laevis S | 1139955720 | 11850000 | 1128105720 | 1.04 | 98.96 |
| X. tropicalis | 1272999256 | 3452000 | 1269547256 | 0.27 | 99.73 |
| tte×ls hybrid #1 | |||||
| Sub genome | Total (bp) | Lost (bp) | Remaining (bp) | Lost (%) | Remaining (%) |
| X. laevis L | 1368982762 | 1084660643 | 284322119 | 79.23 | 20.77 |
| X. laevis S | 1139955720 | 946048452 | 193907268 | 82.99 | 17.01 |
| X. tropicalis | 1272999256 | 5686000 | 1267313256 | 0.45 | 99.55 |
| tte×ls hybrid #2 | |||||
| Sub genome | Total (bp) | Lost (bp) | Remaining (bp) | Lost (%) | Remaining (%) |
| X. laevis L | 1368982762 | 1259268028 | 109714734 | 91.99 | 8.01 |
| X. laevis S | 1139955720 | 1003939197 | 136016523 | 88.07 | 11.93 |
| X tropicalis | 1272999256 | 2964000 | 1270035256 | 0.23 | 99.77 |
| tte×ls hybrid #3 | |||||
| Sub genome | Total (bp) | Lost (bp) | Remaining (bp) | Lost (%) | Remaining (%) |
| X. laevis L | 1368982762 | 1360054762 | 8928000 | 99.35 | 0.65 |
| X laevis S | 1139955720 | 1131571720 | 8384000 | 99.26 | 0.74 |
| X tropicalis | 1272999256 | 3728000 | 1269271256 | 0.29 | 99.71 |
| tte×ls hybrid #4 | |||||
| Sub genome | Total (bp) | Lost (bp) | Remaining (bp) | Lost (%) | Remaining (%) |
| X. laevis L | 1368982762 | 1361764762 | 7218000 | 99.47 | 0.53 |
| X laevis S | 1139955720 | 1134337720 | 5618000 | 99.51 | 0.49 |
| X. tropicalis | 1272999256 | 3240000 | 1269759256 | 0.25 | 99.75 |
| te×bs hybrid | |||||
| Sub genome | Total (bp) | Lost (bp) | Remaining (bp) | Lost (%) | Remaining (%) |
| X. borealis L | 1428994000 | 108866000 | 1320128000 | 7.62% | 92.38% |
| X. borealis S | 1201786000 | 30804000 | 1170982000 | 2.56% | 97.44% |
| X tropicalis | 1273010000 | 11592000 | 1261418000 | 0.91% | 99.09% |
Extended Data Table 4. Overrepresentation test of all or metabolism-only 3L and 4L lost genes.
PANTHER software (http://pantherdb.org/) was used to perform a statistical overrepresentation test on all (top table) or metabolism-only (bottom table) lost genes from chromosomes 3L and 4L. Only overrepresented processes are shown in the top table (*). Only the top 5 processes based on fold enrichment are shown in the bottom table (†).
| 3L and 4L lost genes overrepresentation test | ||||||
|
| ||||||
| Analysis Type | PANTHER Overrepresentation Test (release 20170413) | |||||
| Annotation Version and Release Date | PANTHER version 11.1 Released 2016-10-24 | |||||
| Analyzed List | Client Text Box Input (Xenopus tropicalis) | |||||
| Reference List | Xenopus tropicalis (all genes in database) | |||||
| Bonferroni correction | TRUE | |||||
|
| ||||||
| PANTHER GO-Slim Biological Process* | Xenopus tropicalis - REFLIST (18238) | Client Text Box Input (843) | Client Text Box Input (expected) | Client Text Box Input (fold Enrichment) | Client Text Box Input (P-value) | 95% Confidence Interval (binomial test) |
|
| ||||||
| biosynthetic process (GO:0009058) | 1295 | 141 | 100.05 | 1.41 | 8.01E-03 | [123.4, ∞] |
| nitrogen compound metabolic process (GO:0006807) | 1738 | 179 | 134.27 | 1.33 | 1.40E-02 | [159.6, ∞] |
| metabolic process (GO:0008152) | 6036 | 546 | 466.32 | 1.17 | 1.13E-03 | [522.4, ∞] |
|
| ||||||
| 3L and 4L lost metabolism genes overrepresentation test | ||||||
|
| ||||||
| Analysis Type | PANTHER Overrepresentation Test (release 20170413) | |||||
| Annotation Version and Release Date | PANTHER version 11.1 Released 2016-10-24 | |||||
| Analyzed List | Client Text Box Input (Xenopus tropicalis) | |||||
| Reference List | Xenopus tropicalis (all genes in database) | |||||
| Bonferroni correction | TRUE | |||||
|
| ||||||
| PANTHER GO-Slim Biological Process† | Xenopus tropicalis - REFLIST (18238) | Client Text Box Input (843) | Client Text Box Input (expected) | Client Text Box Input (fold Enrichment) | Client Text Box Input (P-value) | 95% Confidence Interval (binomial test) |
|
| ||||||
| glycolysis (GO:0006096) | 26 | 6 | 0.78 | 7.71 | 3.71E-02 | [2.6, ∞] |
| rRNA metabolic process (GO:0016072) | 104 | 15 | 3.11 | 4.82 | 2.22E-04 | [9.3, ∞] |
| DNA replication (GO: 0006260) | 114 | 16 | 3.41 | 4.69 | 1.38E-04 | [10.1, ∞] |
| tRNA metabolic process (GO:0006399) | 104 | 14 | 3.11 | 4.5 | 1.11E-03 | [8.5, ∞] |
| generation of precursor metabolites and energy (GO:0006091) | 185 | 24 | 5.54 | 4.33 | 9.87E-07 | [16.6, ∞] |
Supplementary Material
Acknowledgments
We thank members of the Heald lab, present and past, for support and fruitful discussions. We thank the students who helped with some of the experiments, Brian Castellano, Jingxun Chen, Stephan Ramos, Armbien Sabillo, and Karen Shih. We are grateful to the Marine Biological Laboratory (MBL) and the National Xenopus Resource (NXR), also for organizing the 2013 Advanced Imaging in Xenopus Workshop where several techniques used here were taught to RG, and to John Wallingford and Asako Shindo for subsequent support. We thank the Welch, King, Harland, Rokhsar, Barton, and Fletcher labs at UC Berkeley for sharing reagents, materials and expertise as well as Todd Stukenberg (University of Virginia) and Aaron Straight (Stanford University) for providing us with the Ndc80 and CENP-A antibodies, respectively. Special thanks to Austin Mudd and Daniel Rokhsar for providing early access to the X. borealis genome assembly. This work used the Functional Genomics Laboratory, a QB3-Berkeley Core Research Facility at UC Berkeley as well as the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant. The confocal microscopy performed in this work was done at the UC Berkeley CRL Molecular Imaging Center, supported by NSF DBI-1041078. RG was initially supported by an EMBO long term fellowship ALTF 836-2013 and for most of this project by an HFSP long term fellowship LT 0004252014-L. RA was supported in part by an NSF REU Summer Fellowship in 2014. RH was supported by NIH R35 GM118183 and the Flora Lamson Hewlett Chair. DKN was supported by NIH R01 CA172667. MK was supported by the UC Berkeley MCB department NIH training grant 4T32GM007232-40. TK was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2016R1C1B2009302), and the UNIST Research Fund (Grant Number 1.160060.01). GJCV, IVK and GG were supported by R01HD069344 (NICHD).
Footnotes
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
RH and RG designed the project. RG performed the molecular, cell and developmental biology experiments, aided by RA, and analyzed the data. MK, together with RG, performed the experiments related to X. borealis and analyzed the data. GJCV, IVK and GG prepared and analyzed the hybrid genomes. BM and DKN performed the metabolomic profiling of hybrids. TK and EMM contributed to the transcriptome data analysis. RG prepared the figures and wrote the manuscript with RH, incorporating feedback from all authors.
References
- 1.Seehausen O, et al. Genomics and the origin of species. Nat Rev Genet. 2014;15:176–192. doi: 10.1038/nrg3644. [DOI] [PubMed] [Google Scholar]
- 2.Presgraves DC. The molecular evolutionary basis of species formation. Nat Rev Genet. 2010;11:175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- 3.Session AM, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:1–15. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown KS, et al. Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J Cell Biol. 2007;176:765–70. doi: 10.1083/jcb.200610043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch N, Zimmerman LB, Grainger RM. Xenopus, the next generation: X. tropicalis genetics and genomics. Dev Dyn. 2002;225:422–33. doi: 10.1002/dvdy.10178. [DOI] [PubMed] [Google Scholar]
- 6.Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–96. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bürki E. The expression of creatine kinase isozymes in Xenopus tropicalis, Xenopus laevis laevis, and their viable hybrid. Biochem Genet. 1985;23:73–88. doi: 10.1007/BF00499114. [DOI] [PubMed] [Google Scholar]
- 8.Narbonne P, Simpson DE, Gurdon JB. Deficient induction response in a Xenopus nucleocytoplasmic hybrid. PLoS Biol. 2011;9:e1001197. doi: 10.1371/journal.pbio.1001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamilton L. Androgenic haploids of a toad, Xenopus laevis. Nature. 1957;179:159. doi: 10.1038/179159a0. [DOI] [PubMed] [Google Scholar]
- 10.Goda T, et al. Genetic screens for mutations affecting development of Xenopus tropicalis. PLoS Genet. 2006;2:e91. doi: 10.1371/journal.pgen.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wühr M, et al. Evidence for an upper limit to mitotic spindle length. Curr Biol. 2008;18:1256–61. doi: 10.1016/j.cub.2008.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheeseman IM. The Kinetochore. Cold Spring Harb Perspect Biol. 2014;6:77–105. doi: 10.1101/cshperspect.a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujiwara a, Abe S, Yamaha E, Yamazaki F, Yoshida MC. Uniparental chromosome elimination in the early embryogenesis of the inviable salmonid hybrids between masu salmon female and rainbow trout male. Chromosoma. 1997;106:44–52. doi: 10.1007/s004120050223. [DOI] [PubMed] [Google Scholar]
- 14.Sakai C, et al. Chromosome elimination in the interspecific hybrid medaka between Oryzias latipes and O. hubbsi. Chromosome Res. 2007;15:697–709. doi: 10.1007/s10577-007-1155-9. [DOI] [PubMed] [Google Scholar]
- 15.Ferree PM, Barbash Da. Species-Specific Heterochromatin Prevents Mitotic Chromosome Segregation to Cause Hybrid Lethality in Drosophila. PLoS Biol. 2009;7:e1000234. doi: 10.1371/journal.pbio.1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalitsis P, Choo KHA. The evolutionary life cycle of the resilient centromere. Chromosoma. 2012;121:327–40. doi: 10.1007/s00412-012-0369-6. [DOI] [PubMed] [Google Scholar]
- 17.Crasta K, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–8. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terradas M, Martín M, Tusell L, Genescà A. DNA lesions sequestered in micronuclei induce a local defective-damage response. DNA Repair (Amst) 2009;8:1225–34. doi: 10.1016/j.dnarep.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Hensey C, Gautier J. A developmental timer that regulates apoptosis at the onset of gastrulation. Mech Dev. 1997;69:183–195. doi: 10.1016/s0925-4773(97)00191-3. [DOI] [PubMed] [Google Scholar]
- 21.Vastag L, et al. Remodeling of the metabolome during early frog development. PLoS One. 2011;6:e16881. doi: 10.1371/journal.pone.0016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma H, et al. Incompatibility between Nuclear and Mitochondrial Genomes Contributes to an Interspecies Reproductive Barrier. Cell Metab. 2016;24:283–294. doi: 10.1016/j.cmet.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HY, et al. Incompatibility of Nuclear and Mitochondrial Genomes Causes Hybrid Sterility between Two Yeast Species. Cell. 2008;135:1065–1073. doi: 10.1016/j.cell.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 24.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: Expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid M, Steinlein C. Chromosome Banding in Amphibia. XXXII. the Genus Xenopus (Anura, Pipidae) Cytogenet Genome Res. 2015;145:201–217. doi: 10.1159/000433481. [DOI] [PubMed] [Google Scholar]
- 26.Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) Garland Publishing; 1994. [Google Scholar]
- 27.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee C, Kieserman E, Gray RS, Park TJ, Wallingford J. Whole-mount fluorescence immunocytochemistry on Xenopus embryos. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot4957. pdb.prot4957. [DOI] [PubMed] [Google Scholar]
- 29.Levy DL, Heald R. Nuclear size is regulated by importin α and Ntf2 in Xenopus. Cell. 2010;143:288–98. doi: 10.1016/j.cell.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maresca TJ, Heald R. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol Biol. 2006;322:459–74. doi: 10.1007/978-1-59745-000-3_33. [DOI] [PubMed] [Google Scholar]
- 31.Murray aW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 32.Hannak E, Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat Protoc. 2006;1:2305–14. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- 33.Edelstein AD, et al. Advanced methods of microscope control using μManager software. J Biol Methods. 2014;1:10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Louie SM, et al. GSTP1 Is a Driver of Triple-Negative Breast Cancer Cell Metabolism and Pathogenicity. Cell Chem Biol. 2016;23:567–78. doi: 10.1016/j.chembiol.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic and transcriptomic data generated for this study are available from public databases: stage 9 te×ls hybrid whole genome sequencing data (NCBI SRA: SRP124316; https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP124316) and corresponding stage 9 X. laevis, X. tropicalis and le×ts hybrid controls (NCBI GEO: GSE92382; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE92382), tailbud and tadpole stage tte×ls hybrid whole genome sequencing data (NCBI SRA: SRP124316; https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP124316), stage 9 te×bs hybrid whole genome sequencing data (NCBI SRA: SRP124316; https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP124316) and 7 hpf X. tropicalis and te×ls hybrid transcriptome RNA-seq data (NCBI GEO: GSE106157; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE106157).