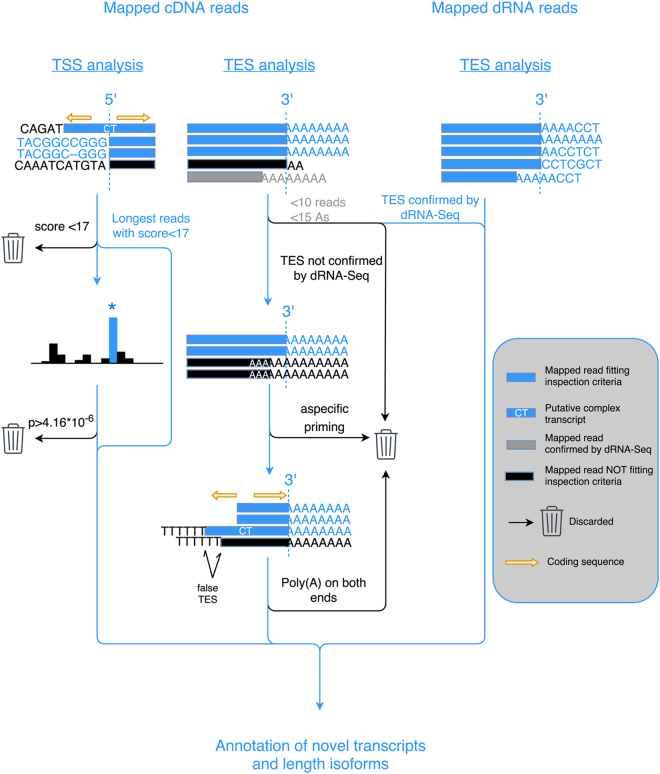
Figure 1.
The schematic representation of the workflow used to annotate novel transcripts. Only the cDNA-sequencing reads were used for identifying TSSs. The read start distribution was obtained by only counting reads which contained the 5′ adaptor sequence. Nucleotide positions where the read start distribution was significantly different from a Poisson distribution were annotated as TSSs. In the case of complex transcripts, if no TSS could be determined, the closest upstream TSS was assumed to be the TSS. Both dRNA and cDNA reads were used to identify TESs. In case of the dRNA-Seq, support by one polyA-containing read was sufficient to annotate a TES, while in the case of cDNA-Seq, at least 10 reads were required to support a given TES, and also reads that could have arisen through non-specific priming, and reads containing poly(A) sequences on both ends were discarded from the TES analysis. The complex transcripts, supported by reads with double polyA-tails, were annotated, and their orientation was determined according to the polyA-tail, which passed the TES annotation criteria.