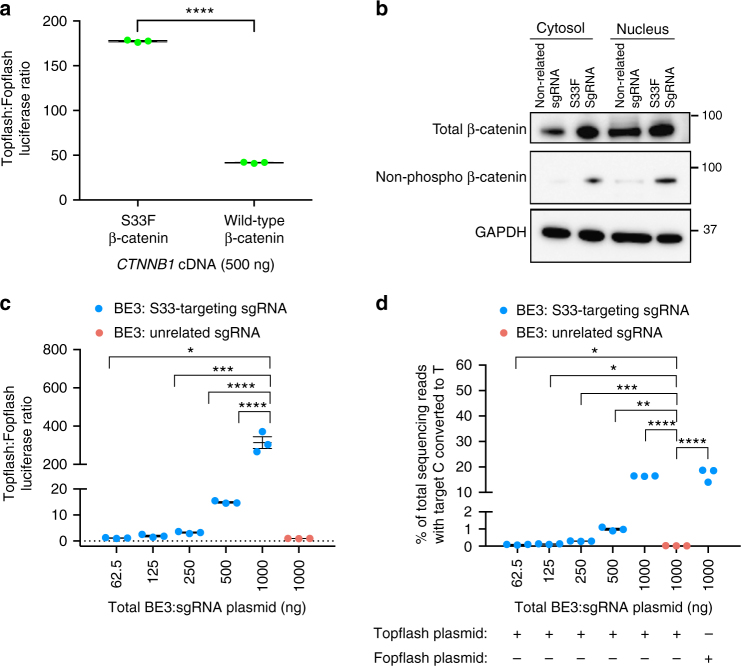
Fig. 2.
Biological outcomes associated with base editing S33F in β-catenin in human cells. a HEK293T cells were transfected with plasmids encoding Topflash (β-catenin-responsive firefly luciferase reporter) or Fopflash (mutant form of Topflash that cannot be activated by β-catenin) and mutant S33F β-catenin or wild-type (Ser 33) β-catenin. Wnt signaling was measured by the ratio of Topflash:Fopflash luciferase activity. b Cytosolic and nuclear extracts of HEK293T cells treated with base editor and S33-targeting sgRNA or unrelated sgRNA were subjected to western blot analysis for total β-catenin or non-phosphorylated β-catenin (Ser33/Ser37/Thr41). Each blot represents one antibody. GAPDH was used as a loading control. c HEK293T cells were transfected with plasmids encoding the Topflash or Fopflash reporters, base editor, and S33F or unrelated control sgRNA. The Topflash:Fopflash luciferase ratio for BE3 + S33-targeting sgRNA (blue) and BE3 + unrelated sgRNA (red) are shown. d Percent C-to-T conversion at the target Ser 33 codon, which results in the S33F mutation in β-catenin, assayed by HTS. Values and error bars reflect mean ± S.E.M. of three biological replicates performed on different days. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, and **** p ≤ 0.0001 (Student’s two tailed t-test)