Abstract
Chronic hepatitis B virus infection is a significant risk factor for cirrhosis and hepatocellular carcinoma. The HBx protein is required for virus replication, but the lack of robust infection models has hindered our understanding of HBx functions that could be targeted for antiviral purposes. We briefly review three properties of HBx: its binding to DDB1 and its regulation of cell survival and metabolism, to illustrate how a single viral protein can have multiple effects in a cell. We propose that different functions of HBx are needed, depending on the changing hepatocyte environment encountered during a chronic virus infection, and that these functions might serve as novel therapeutic targets for inhibiting hepatitis B virus replication and the development of associated diseases.
Graphical abstract
Introduction
The human hepatitis B virus (HBV) is a major human pathogen. An HBV infection can be acute or chronic, with the latter affecting over 240 million patients worldwide and leading to cycles of liver inflammation and significant deaths from liver failure and hepatocellular carcinoma (HCC) ([1] and reviewed in [2]). The HBV lifecycle is complex (Fig. 1) (reviewed in [3,4]). Upon entering hepatocytes, the partially double-stranded virion DNA genome is converted into viral covalently closed circular DNA (cccDNA), which serves as the transcriptional template. cccDNA is very stable, is considered to be a cause of viral persistence, and is one target of the HBV regulatory HBx protein (reviewed in [5]). The purpose of this brief review is to summarize functions of the HBV HBx protein that might contribute to maintenance of a persistent HBV infection and could therefore be potential therapeutic targets for the interruption of chronic HBV replication. Although numerous HBx activities that could affect persistent HBV replication have been reported, we focus on three HBx functions. We apologize to colleagues who have defined other HBx activities that might also be important for persistent HBV replication but could not be described due to space limitations.
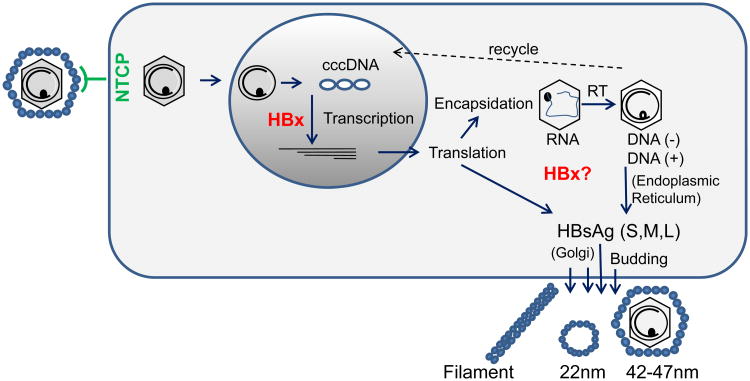
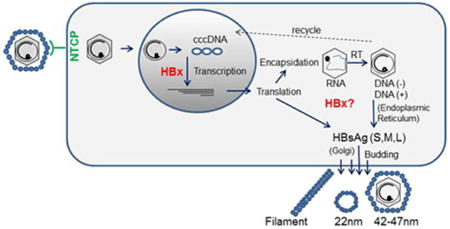
Figure 1.
HBV lifecycle. Virus particles containing partially double-stranded (ds) DNA (∼dsDNA) genomes enter the cell via the NTCP receptor. Following uncoating of surface antigen (small blue circles), the core particles (hexagons) deliver the genome to the nucleus. The ∼dsDNA is repaired by host factors and converted into covalently closed circular (ccc)DNA. The cccDNA serves as the template for HBx-mediated viral transcription. The viral mRNAs (shown in the nucleus) are transported to the cytoplasm for translation. The 3.5-kb pregenomic RNA and a copy of the viral polymerase (small black circles) is encapsidated and reverse transcribed (RT) into the negative-strand DNA, which is then copied into positive-strand DNA. Viral cores move through the endoplasmic reticulum and Golgi, where they acquire surface antigen (envelope) and bud from the cell. Cytoplasmic-core particles may alternatively recycle back to the nucleus.
Natural history of chronic HBV
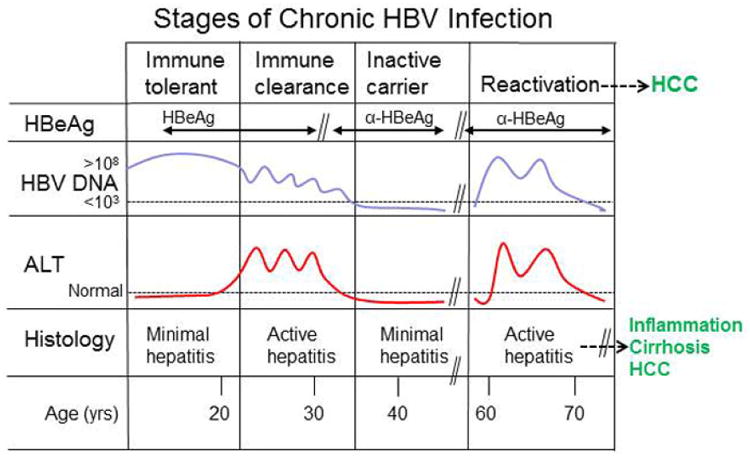
Chronic HBV infection is thought to occur in four sequential stages that can be defined by specific serum markers and histological examination of liver tissue [4,6] (Fig. 2). The first stage, immune tolerance, is characterized by high-titer HBV DNA, expression of the HBV HBeAg, a marker of active HBV replication, and normal levels of alanine aminotransferase (ALT), a marker of potential liver damage. Liver tissue shows mild to no inflammatory changes, although events contributing to cirrhosis and HCC may still be occurring during this stage (reviewed in [7]). The second stage, immune clearance, features variable and declining levels of HBV DNA, concomitant spikes in ALT levels, and active liver inflammation (hepatitis). There may also be a conversion from HBeAg-positivity to anti-HBeAg-positivity. The third stage, the inactive carrier stage, is marked by the presence of anti-HBeAg positivity, low-to-undetectable HBV DNA, normal ALT levels, and a return to minimal hepatitis. In the fourth or reactivation stage, there are again spikes of HBV replication, increased ALT, and active hepatitis. Repeated cycles of reactivation and inflammation may lead to cirrhosis and HCC. Chronic HBV infection lasts for decades, and the virus-host interactions underlying progression through various stages of the infection remain incompletely understood. The HBV HBx protein is presumed to be expressed throughout chronic HBV infection based on detection of the analogous WHx protein in woodchucks chronically infected with the woodchuck hepatitis virus (WHV), a member of the same virus family as HBV [8]. HBx likely has multiple functions that could vary depending on the specific stage of chronic infection and the cellular factors encountered by the virus. These functions may be reflected in the numerous activities that have been ascribed to HBx in different experimental models.
Figure 2.
Four stages of a chronic HBV infection. Chronic HBV infection typically proceeds through four stages, .as described in the text. We propose that HBx activities may differ depending on the cellular factors present during the different stages of a chronic infection.
HBx and virus replication
The HBV genome encodes four overlapping open-reading frames (ORFs) including the X ORF that encodes HBx. HBx is required to initiate and maintain HBV replication in HepaRG cells [9] and human-liver-chimeric mice [10], and WHx is required for WHV replication in woodchucks [11,12]. In plasmid-transient-transfection assays with a greater-than-unit length HBV, or a similar HBV lacking HBx expression, HBx is required for maximal virus replication [13–15]. HBx localizes to both a Triton X-100 detergent-soluble and insoluble (cytoskeletal) fraction, where its half-life is 15-30 minutes or 3 hours, respectively [16–19]. In the nucleus, HBx interacts with cccDNA [20] and basal transcriptional machinery and activates transcription (reviewed in [21–23]). In the cytoplasm, HBx stimulates signal transduction pathways to benefit virus replication, including factors that affect cell survival, metabolism, proliferation, and transcription pathways (reviewed in [21–24]). A fraction of cytosolic HBx localizes to the outer mitochondrial membrane and interacts with the voltage-dependent anion channel [25]. Technical considerations for working with HBx, as well as comprehensive reviews of HBx activities, have been published [21–23,26,27]. Here we review a few more recent findings of HBx activities that are likely of relevance during chronic HBV infection.
HBx and DDB1
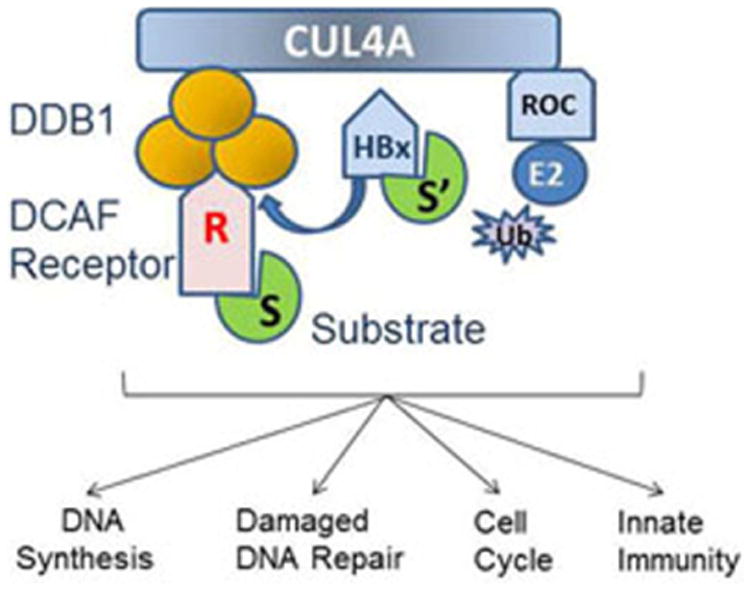
Viruses with limited genetic information frequently usurp cellular pathways to facilitate their replication and interact with cellular proteins to mediate their role(s) in viral replication. It is clear that the binding of HBx to damaged DNA binding protein 1 (DDB1) [28,29] is critical for virus replication in both woodchucks [30] and in the HBV-plasmid, HepG2-replication model [31,32]. DDB1 was originally identified as a cofactor in the recognition step of nucleotide-excision repair (NER) [33]. More recently, DDB1 was shown to be an adaptor protein for the Cullin 4A RING E3 Ligase (CRL4) and acts by binding DDB1 Cullin Accessory Factor (DCAF) receptors that recruit substrate proteins for ubiquitination and degradation [34,35] (Fig. 3). In this way, CRL4 regulates diverse cellular processes such as damaged-DNA repair (DDR), the cell cycle, and innate immunity (reviewed in [36]). Of significance, HBx is a viral DCAF receptor [32,37] and is proposed to impact downstream DDB1-DCAF pathways (Fig. 3).
Figure 3.
HBx and DDB1. DDB1 (orange) is an adaptor protein of the Cullin 4A-DDB1 E3 Ligase (CRL4) and acts by recruiting DCAF-receptor proteins (R) that bind substrates (S) that are ubiquitinated (Ub) and degraded to regulate downstream pathways such as DNA synthesis, damaged-DNA repair, the cell cycle, and innate immunity. HBx is a viral DCAF and binds DDB1 as a required step in virus replication. HBx-DDB1 may recruit new substrates (S′) to the CRL4 or may alter downstream pathways regulated by CRL4. Other key proteins of the complex are the RING protein (Roc) that binds to the E2 enzyme.
Activation of the DDR by viruses is a common strategy to provide factors needed for virus replication (reviewed in [38,39]). HBV (or HBx) can both activate and inhibit DDR pathways. Incubation of human liver HL7702 cells with HBV-positive serum activated (phosphorylated) Ataxia telangiectasia-mutated (ATM)-Rad3-related (ATR) [40]. A similar activation of ATR was reported in HBx-inducible, immortalized-murine hepatocytes cultured in low serum [41,42]. HBx also stimulates the DNA helicase activity of the TFIIH subunits [43] and utilizes the host DNA-repair enzyme TDP2 when synthesizing viral cccDNA [44]. Some studies, however, support the idea that HBx inhibits the NER portion of the DDR. HBx inhibits NER in HepG2 cells [45–47] and in primary mouse hepatocytes [48,49]. It also inhibits base-excision repair [50]. During chronic HBV infection, HBx may need to both activate and inhibit DDR, depending on the hepatocyte environment and the specific DDR function needed to benefit that stage of virus replication. Existing chemical inhibitors of ATR, and the related ataxia telangiectasia-mutated (ATM) pathways of DDR, are potential antiviral agents [51], but more studies of HBx function in authentic HBV-infection models are needed.
Several viruses target CRL4 and recruit cellular substrates whose degradation benefits virus replication (reviewed in [52–54]). An early study concluded that HBx inhibits proteasome function to preserve a factor needed by the virus [55]. More recently, an opposing view has emerged from studies showing that HBx binds to the cellular DDB1-containing E3 ligase and promotes the degradation of the structural maintenance of chromosomes (Smc) complex Smc5/6 [56,57]. Smc5/6 normally inhibits transcription from the HBV cccDNA template, and degradation of Smc5/6 eases the transcriptional repression, leading to increased viral mRNA synthesis (reviewed in [58]). This function of HBx requires co-localization with Nuclear Domain 10 (ND10) [59]. Other HBV restriction factors are under investigation (Table 1), some of which may also be targeted for degradation, although the mechanism(s) by which this occurs may differ (see Table 1 and references therein). Since HBx binds over 100 cellular proteins [68], it is likely that additional HBV restriction factors will be discovered. The presence of antiviral restriction factors is predicted to vary at different times during a chronic virus infection and could act at any stage of the virus lifecycle.
Table 1. Putative HBV restriction factors.
| Protein | Step of HBV replication inhibited | Reference |
|---|---|---|
| Apobec3g | Release from plasma membrane | [56,57] |
| Smc6 | Transcription from cccDNA | [58,59] |
| Samhd1 | Viral DNA synthesis | [60,61] |
| Tln1 | Transcription from cccDNA | [62] |
| Ddx3 | Post-encapsidation of pgRNA | [63] |
| Ddx5 | Transcription from cccDNA | [64] |
| Zeb2 | Transcription from cccDNA | [65] |
Many viruses encode proteins that function, in part, by deregulating cell-cycle checkpoints to benefit virus replication (reviewed in [69,70]). HBx can induce cells from G0 to G1, but then cause cells to stall at the G1/S border (reviewed in [22,24,71]). The CRL4 associated with DCAF receptor CDT2 (CRL4CDT2) has been implicated in cell-cycle progression (reviewed in [72]), and this is accomplished via the degradation of p21/CIP/WAF1 and of DNA-replication-licensing factor Cdt1 (reviewed in [73]). Interestingly, p21 levels are reduced in the regenerating liver of HBx-transgenic mice [74], and HBx promotes DNA re-replication in immortalized murine hepatocytes [42] but not in HeLa cells [75]. Thus, the ability of HBx to promote and/or inhibit cell-cycle progression may be context specific and important during different stages of chronic infection.
HBx and cell survival
HBx has been reported to activate, inhibit, or have no effect on cellular apoptosis pathways; some studies have also suggested that HBx may sensitize cells to other factors that regulate cell survival (reviewed in [76,77]). These seemingly contradictory observations probably reflect cell-specific consequences of HBx expression and are likely to be relevant to differing HBx effects at various steps of the viral lifecycle or different stages of a chronic HBV infection (Fig. 2). While many early studies of HBx regulation of apoptosis were conducted in immortalized or transformed cells, more recent studies in ex vivo models of primary hepatocyte systems have shown that HBx has a dual role in regulating apoptosis [78,79]. HBx was shown to be anti-apoptotic in these systems via activation of the transcription factor NFkB, an activator of anti-apoptotic signals [78]. In contrast, when NFκB was inhibited, HBx was pro-apoptotic. Additional studies in primary hepatocytes showed that HBx also activates the anti-apoptotic factor AKT; this HBx activity promoted hepatocyte survival at the expense of high levels of HBV replication [79]. Inhibition of AKT stimulated HBV replication but also led to HBx-induced apoptosis. These observations are also consistent with data showing that HBx sensitizes cells to apoptotic signals [80–83] and suggest that HBx pro-apoptotic effects are normally masked by its activation of NF-κB or AKT but could become apparent when other pro-apoptotic signals are present. Studies in HBV- and HBx-transgenic mice and in liver-derived cell lines suggest that HBV and HBx elevate expression of pro-apoptotic BAX and lower expression of anti-apoptotic Bcl-xL, which could sensitize hepatocytes to pro-apoptotic signals while not directly inducing apoptosis [83,84]. HBx localization to mitochondria, a cellular signaling hub that controls cell survival, might also affect apoptotic signals [25,78,85–88]. The results of one study showed that HBx expression elevated reactive oxygen species (ROS) levels, which typically come from mitochondria, decreased expression of mitochondrial oxidative phosphorylation enzymes, and sensitized cells to pro-apoptotic signals [81]. HBV and HBx also induced mitophagy in liver-derived cells, and inhibition of mitophagy in these cells induced apoptosis [86]. In this scenario, activation of mitophagy served to protect cells. Overall, the results of recent studies suggest that noncytopathic, persistent HBV replication is linked to HBx activation of anti-apoptotic signals that also lower the levels of HBV replication; when HBx anti-apoptotic signals are blocked, HBV replication is elevated but HBx pro-apoptotic signals kill the infected cells [79]. Whether HBx pro- or anti-apoptotic activities influence specific steps of the HBV lifecycle, or specific stages of an HBV infection, remains to be determined.
HBx and metabolism
The possible role for HBx-mediated alterations in hepatocyte metabolism, and the consequences for persistent HBV replication, is an emerging area of investigation [89]. HBx-mediated changes in hepatocyte metabolism have been analyzed in mouse livers, ex vivo hepatocyte culture systems, and liver-derived cell lines when HBx was expressed alone and in the context of HBV replication [90,91]. These studies suggest that HBx can elevate the expression of hepatic gluconeogenic factors, such as peroxisome proliferator-activated receptor gamma coactivator-1α and phosphoenolpyruvate carboxykinase, and can activate major metabolic signaling pathways including the PI3K/AKT, mammalian target of rapamycin complex 1 (mTORC1), and AMP-activated protein kinase (AMPK) pathways [90,91]. These HBx effects have been linked to control of HBV transcription and genome replication. Interestingly, in primary hepatocyte systems, HBx simultaneously activated the opposing mTORC1- and AMPK-signaling pathways. Activation of mTORC1 inhibited HBV replication, and activation of AMPK enhanced HBV replication [91]. The results of a study that analyzed PI3K/AKT and mTORC1 signaling in HBV-positive patient liver samples also demonstrated that mTORC1 and AKT levels were elevated [92]. The cumulative consequences of HBx-induced alterations of metabolic signaling pathways may be to provide lipids, proteins, and nucleotides required for HBV replication while also altering normal hepatocyte metabolism so as to promote or contribute to progression of HBV-associated carcinogenesis.
Conclusions
Given the clinical importance of chronic HBV infection and liver disease, and the central role of HBx in HBV replication, it is important to consider the idea of targeting HBx to interrupt virus replication. We propose that HBx serves multiple functions during the various stages of chronic HBV infection. However, a recurring theme for known HBx-host interactions is that HBx can both promote and inhibit cellular pathways with which it interacts. HBx may need this flexibility in order to mediate its function(s) in the different cellular environments encountered during a decades-long chronic HBV infection. At present, it is challenging to identify a specific function of HBx that might be targeted. Moving forward, more robust HBV-infection models are needed in order to study HBx functions relevant to virus replication in different physiological settings.
Highlights.
HBV HBx is required for virus replication.
Required HBx function(s) may vary with the stage of chronic HBV infection.
HBx binding to DDB1 may impact several DDB1-regulated pathways.
HBx effects on apoptosis and metabolism are likely vital for chronic HBV infection.
DDB1 and cell survival and metabolism pathways are potential anti-HBV targets.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.World Health Organization (WHO) Hepatitis B: Fact Sheet. 2017 Available from http:/www.who-int/mediacentre/factsheets.
- 2.Fallot G, Neuveut C, Buendia MA. Diverse roles of hepatitis B virus in liver cancer. Curr Opin Virol. 2012;2:467–473. doi: 10.1016/j.coviro.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Seeger C, Zoulim F, Mason WS. Hepadnaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Sixth. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 2185–2221. [Google Scholar]
- 4.Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;479-480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 6.Yim HJ, Lok AS. Natural history of chronic hepatitis B virus infection: what we knew in 1981 and what we know in 2005. Hepatology. 2006;43:S173–S181. doi: 10.1002/hep.20956. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy PTF, Litwin S, Dolman GE, Bertoletti A, Mason WS. Immune Tolerant Chronic Hepatitis B: The Unrecognized Risks. Viruses. 2017;9:1–19. doi: 10.3390/v9050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dandri M, Schirmacher P, Rogler CE. Woodchuck hepatitis virus X protein is present in chronically infected woodchuck liver and woodchuck hepatocellular carcinomas which are permissive for viral replication. J Virol. 1996;70:5246–5254. doi: 10.1128/jvi.70.8.5246-5254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Lucifora J, Arzberger S, Durantel D, Belloni L, Strubin M, Levrero M, Zoulim F, Hantz O, Protzer U. Hepatitis B Virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol. 2011;55:996–1003. doi: 10.1016/j.jhep.2011.02.015. Demonstrated that HBx is required for HBV infection of HepaRG cells, but not for the generation of cccDNA. [DOI] [PubMed] [Google Scholar]
- 10••.Tsuge M, Hiraga N, Akiyama R, Tanaka S, Matsushita M, Mitsui F, Abe H, Kitamura S, Hatakeyama T, Kimura T, Miki D, Mori N, Imamura M, Takahashi S, Hayes CN, Chayama K. HBx protein is indispensable for development of viraemia in human hepatocyte chimeric mice. J Gen Virol. 2010;91:1854–1864. doi: 10.1099/vir.0.019224-0. Showed that HBx is required for viremia in human liver chimeric mouse model. [DOI] [PubMed] [Google Scholar]
- 11.Chen HS, Kaneko S, Girones R, Anderson RW, Hornbuckle WE, Tennant BC, Cote PJ, Gerin JL, Purcell RH, Miller RH. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scaglioni PP, Melegari M, Wands JR. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J Virol. 1997;71:345–353. doi: 10.1128/jvi.71.1.345-353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melegari M, Scaglioni PP, Wands JR. Cloning and characterization of a novel hepatitis B virus X binding protein that inhibits viral replication. J Virol. 1998;72:1737–1743. doi: 10.1128/jvi.72.3.1737-1743.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2002;294:2376–2378. doi: 10.1126/science.294.5550.2376. First demonstration that HBx could complement HBx-deficient replication in HBV plasmid replication model. [DOI] [PubMed] [Google Scholar]
- 16.Henkler F, Hoare J, Waseem N, Goldin RD, McGarvey MJ, Koshy R, King IA. Intracellular localization of the hepatitis B virus HBx protein. J Gen Virol. 2001;82:871–882. doi: 10.1099/0022-1317-82-4-871. [DOI] [PubMed] [Google Scholar]
- 17.Hoare J, Henkler F, Dowling JJ, Errington W, Goldin RD, Fish D, McGarvey MJ. Subcellular localization of the X protein in HBV infected hepatocytes. J Med Virol. 2001;64:419–426. doi: 10.1002/jmv.1067. [DOI] [PubMed] [Google Scholar]
- 18.Schek N, Bartenschlager R, Kuhn C, Schaller H. Phosphorylation and rapid turnover of hepatitis B virus X-protein expressed in HepG2 cells from a recombinant vaccinia virus. Oncogene. 1991;6:1735–1744. [PubMed] [Google Scholar]
- 19.Dandri M, Petersen J, Stockert RJ, Harris TM, Rogler CE. Metabolic labeling of woodchuck hepatitis B virus X protein in naturally infected hepatocytes reveals a bimodal half-life and association with the nuclear framework. J Virol. 1998;72:9359–9364. doi: 10.1128/jvi.72.11.9359-9364.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci U S A. 2009;106:19975–19979. doi: 10.1073/pnas.0908365106. Demonstrated that HBx can localize to cccDNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benhenda S, Cougot D, Buendia MA, Neuveut C. Hepatitis B virus X protein molecular functions and its role in virus life cycle and pathogenesis. Adv Cancer Res. 2009;103:75–109. doi: 10.1016/S0065-230X(09)03004-8. [DOI] [PubMed] [Google Scholar]
- 23.Lucifora J, Protzer U. Hepatitis B Virus X Protein: A Key Regulator of the Virus Life Cycle. In: Garcia ML, Romanowski V, editors. Viral Genomes- Molecular Structure, Diversity, Gene Expression Mechanisms and Host-Virus Interactions. 2012. pp. 141–154. [Google Scholar]
- 24.Casciano JC, Bagga S, Yang B, Bouchard MJ. Modulation of Cell Proliferation Pathways by the Hepatitis B Virus X Protein: A Potential Contributor to the Development of Hepatocellular Carcinoma. In: Joseph W, Lau Y, editors. Hepatocellular Carcinoma - Basic Research. 2012. pp. 103–152. [Google Scholar]
- 25•.Rahmani Z, Huh KW, Lasher R, Siddiqui A. Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel and alters its transmembrane potential. J Virol. 2000;74:2840–2846. doi: 10.1128/jvi.74.6.2840-2846.2000. First demonstration of a direct interaction between HBx and an outer mitochondrial membrane protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slagle BL, Andrisani OM, Bouchard MJ, Lee CG, Ou JH, Siddiqui A. Technical standards for hepatitis B virus X protein (HBx) research. Hepatology. 2015;61:1416–1424. doi: 10.1002/hep.27360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slagle BL, Bouchard MJ. Hepatitis B Virus X and Regulation of Viral Gene Expression. Cold Spring Harb Perspect Med. 2016 doi: 10.1101/cshperspect.a021402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee TH, Elledge SJ, Butel JS. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sitterlin D, Lee TH, Prigent S, Tiollais P, Butel JS, Transy C. Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J Virol. 1997;71:6194–6199. doi: 10.1128/jvi.71.8.6194-6199.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Sitterlin D, Bergametti F, Tiollais P, Tennant BC, Transy C. Correct binding of viral X protein to UVDDB-p127 cellular protein is critical for efficient infection by hepatitis B viruses. Oncogene. 2000;19:4427–4431. doi: 10.1038/sj.onc.1203770. Demonstrated that HBx-DDB1 is required for virus replication in the woodchuck HBV model in vivo. [DOI] [PubMed] [Google Scholar]
- 31.Leupin O, Bontron S, Schaeffer C, Strubin M. Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death. J Virol. 2005;79:4238–4245. doi: 10.1128/JVI.79.7.4238-4245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodgson AJ, Hyser JM, Keasler VV, Cang Y, Slagle BL. Hepatitis B virus regulatory HBx protein binding to DDB1 is required but is not sufficient for maximal HBV replication. Virol. 2012;426:73–82. doi: 10.1016/j.virol.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abramic M, Levine AS, Protic M. Purification of an ultraviolet-inducible, damage-specific DNA-binding protein from primate cells. J Biol Chem. 1991;266:22493–22500. [PubMed] [Google Scholar]
- 34.Higa LA, Zhang H. Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Division. 2007;2:5–13. doi: 10.1186/1747-1028-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J, Zhou P. DCAFs, the missing link of the Cul4-DDB1 Ubiquitin Ligase. Mol Cell. 2007;26:775–780. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Petroski MD, Deshaies RJ. Function and regulation of Cullin-RING ubiquitin ligases. Nature Reviews Molecular Cell Biology. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 37.Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol. 2010;17:105–111. doi: 10.1038/nsmb.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weitzman MD, Carson CT, Schwartz RA, Lilley CE. Interactions of viruses with the cellular DNA repair machinery. DNA Repair (Amst) 2004;3:1165–1173. doi: 10.1016/j.dnarep.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Hollingworth R, Grand RJ. Modulation of DNA damage and repair pathways by human tumour viruses. Viruses. 2015;7:2542–2591. doi: 10.3390/v7052542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao F, Hou NB, Yang XL, He X, Liu Y, Zhang YH, Wei CW, Song T, Li L, Ma QJ, Zhong H. Ataxia telangiectasia-mutated-Rad3-related DNA damage checkpoint signaling pathway triggered by hepatitis B virus infection. World J Gastroenterol. 2008;14:6163–6170. doi: 10.3748/wjg.14.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang WH, Hullinger RL, Andrisani OM. Hepatitis B virus X protein via the p38MAPK pathway induces E2F1 release and ATR kinase activation mediating p53 apoptosis. J Biol Chem. 2008;283:25455–25467. doi: 10.1074/jbc.M801934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rakotomalala L, Studach L, Wang WH, Gregori G, Hullinger RL, Andrisani O. Hepatitis B virus X protein increases the Cdt1-to-geminin ratio inducing DNA re-replication and polyploidy. J Biol Chem. 2008;283:28729–28740. doi: 10.1074/jbc.M802751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qadri I, Conaway JW, Conaway RC, Schaack J, Siddiqui A. Hepatitis B virus transactivator protein, HBx, associates with the components of TFIIH and stimulates the DNA helicase activity of TFIIH. Proc Natl Acad Sci. 1996;93:10578–10583. doi: 10.1073/pnas.93.20.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koniger C, Wingert I, Marsmann M, Rosler C, Beck J, Nassal M. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc Natl Acad Sci U S A. 2014;111:E4244–E4253. doi: 10.1073/pnas.1409986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Becker SA, Lee TH, Butel JS, Slagle BL. Hepatitis B virus X protein interferes with cellular DNA repair. J Virol. 1998;72:266–272. doi: 10.1128/jvi.72.1.266-272.1998. First report that HBx inhibits NER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groisman IJ, Koshy R, Henkler F, Groopman JD, Alaoui-Jamali MA. Downregulation of DNA excision repair by the hepatitis B virus-x protein occurs in p53-proficient and p53-deficient cells. Carcin. 1999;20:479–483. doi: 10.1093/carcin/20.3.479. [DOI] [PubMed] [Google Scholar]
- 47.Jia L, Wang XW, Harris CC. Hepatitis B virus X protein inhibits nucleotide excision repair. Int J Cancer. 1999;80:875–879. doi: 10.1002/(sici)1097-0215(19990315)80:6<875::aid-ijc13>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 48.Prost S, Ford JM, Taylor C, Doig J, Harrison DJ. Hepatitis B x protein inhibits p53-dependent DNA repair in primary mouse hepatocytes. J Biol Chem. 1998;273:33327–33332. doi: 10.1074/jbc.273.50.33327. [DOI] [PubMed] [Google Scholar]
- 49.Madden CR, Finegold MJ, Slagle BL. Expression of hepatitis B virus X protein does not alter the accumulation of spontaneous mutations in transgenic mice. J Virol. 2000;74:5266–5272. doi: 10.1128/jvi.74.11.5266-5272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van de Klundert MA, van Hemert FJ, Zaaijer HL, Kootstra NA. The hepatitis B virus x protein inhibits thymine DNA glycosylase initiated base excision repair. PLoS One. 2012;7:e48940. doi: 10.1371/journal.pone.0048940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng Z, Li J, Sun J, Song T, Wei C, Zhang Y, Rao G, Chen G, Li D, Yang G, Han B, Wei S, Cao C, Zhong H. Inhibition of HBV replication by theophylline. Antiviral Res. 2011;89:149–155. doi: 10.1016/j.antiviral.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 52.Barry M, Fruh K. Viral modulators of Cullin RING Ubiquitin Ligases: Culling the host defense. Science's stke. 2006;335:1–5. doi: 10.1126/stke.3352006pe21. [DOI] [PubMed] [Google Scholar]
- 53.Minor MM, Slagle BL. Hepatitis B virus HBx protein interactions with the ubiquitin proteasome system. Viruses. 2014;6:4683–4702. doi: 10.3390/v6114683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Exp Cell Res. 2009;315:1610–1618. doi: 10.1016/j.yexcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Z, Torii N, Furusaka A, Malayaman N, Hu ZY, Liang TJ. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J Biol Chem. 2000;275:15157–15165. doi: 10.1074/jbc.M910378199. [DOI] [PubMed] [Google Scholar]
- 56••.Decorsiere A, Mueller H, van Breugel PC, Abdul F, Gerossier L, Beran RK, Livingston CM, Niu C, Fletcher SP, Hantz O, Strubin M. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386–389. doi: 10.1038/nature17170. Showed that HBx recruits HBV restriction factor Smc5/6 to the proteasome for degradation. [DOI] [PubMed] [Google Scholar]
- 57.Murphy CM, Xu Y, Li F, Nio K, Reszka-Blanco N, Li X, Wu Y, Yu Y, Xiong Y, Su L. Hepatitis B Virus X Protein Promotes Degradation of SMC5/6 to Enhance HBV Replication. Cell Rep. 2016;16:2846–2854. doi: 10.1016/j.celrep.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Livingston CM, Ramakrishnan D, Strubin M, Fletcher SP, Beran RK. Identifying and Characterizing Interplay between Hepatitis B Virus X Protein and Smc5/6. Viruses. 2017;9:69. doi: 10.3390/v9040069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niu C, Livingston CM, Li L, Beran RK, Daffis S, Ramakrishnan D, Burdette D, Peiser L, Salas E, Ramos H, Yu M, Cheng G, Strubin M, Delaney WE, IV, Fletcher SP. The Smc5/6 Complex Restricts HBV when Localized to ND10 without Inducing an Innate Immune Response and Is Counteracted by the HBV X Protein Shortly after Infection. PLoS One. 2017;12:e0169648. doi: 10.1371/journal.pone.0169648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 61.Chen R, Zhao X, Wang Y, Xie Y, Liu J. Hepatitis B virus X protein is capable of down-regulating protein level of host antiviral protein APOBEC3G. Sci Rep. 2017(7):40783. doi: 10.1038/srep40783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sommer AF, Riviere L, Qu B, Schott K, Riess M, Ni Y, Shepard C, Schnellbacher E, Finkernagel M, Himmelsbach K, Welzel K, Kettern N, Donnerhak C, Munk C, Flory E, Liese J, Kim B, Urban S, Konig R. Restrictive influence of SAMHD1 on Hepatitis B Virus life cycle. Sci Rep. 2016;6:26616. doi: 10.1038/srep26616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jeong GU, Park IH, Ahn K, Ahn BY. Inhibition of hepatitis B virus replication by a dNTPase-dependent function of the host restriction factor SAMHD1. Virology. 2016;495:71–78. doi: 10.1016/j.virol.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 64.van de Klundert MA, van den Biggelaar M, Kootstra NA, Zaaijer HL. Hepatitis B Virus Protein X Induces Degradation of Talin-1. Viruses. 2016;8:1–17. doi: 10.3390/v8100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ko C, Lee S, Windisch MP, Ryu WS. DDX3 DEAD-box RNA helicase is a host factor that restricts hepatitis B virus replication at the transcriptional level. J Virol. 2014;88:13689–13698. doi: 10.1128/JVI.02035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang H, Xing Z, Mani SK, Bancel B, Durantel D, Zoulim F, Tran EJ, Merle P, Andrisani O. RNA helicase DEAD box protein 5 regulates Polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology. 2016;64:1033–1048. doi: 10.1002/hep.28698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He Q, Li W, Ren J, Huang Y, Huang Y, Hu Q, Chen J, Chen W. ZEB2 inhibits HBV transcription and replication by targeting its core promoter. Oncotarget. 2016;7:16003–16011. doi: 10.18632/oncotarget.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu ZJ, Zhu Y, Huang DR, Wang ZQ. Constructing the HBV-human protein interaction network to understand the relationship between HBV and hepatocellular carcinoma. J Exp Clin Cancer Res. 2010;29:146. doi: 10.1186/1756-9966-29-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bagga S, Bouchard MJ. Cell cycle regulation during viral infection. Methods Mol Biol. 2014;1170:165–227. doi: 10.1007/978-1-4939-0888-2_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Op De Beeck A, Caillet-Fauquet P. Viruses and the cell cycle. Prog Cell Cycle Res. 2001;3:1–19. doi: 10.1007/978-1-4615-5371-7_1. [DOI] [PubMed] [Google Scholar]
- 71.Madden CR, Slagle BL. Stimulation of cellular proliferation by hepatitis B virus X protein. Dis Markers. 2001;17:153–157. doi: 10.1155/2001/571254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee J, Zhou P. Pathogenic Role of the CRL4 Ubiquitin Ligase in Human Disease. Front Oncol. 2012;2:21. doi: 10.3389/fonc.2012.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hannah J, Zhou P. Regulation of DNA damage response pathways by the cullin-RING ubiquitin ligases. DNA Repair (Amst) 2009;8:536–543. doi: 10.1016/j.dnarep.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hodgson AJ, Keasler VV, Slagle BL. Premature cell cycle entry induced by hepatitis B virus regulatory HBx protein during compensatory liver regeneration. Canc Res. 2008;68:10341–10348. doi: 10.1158/0008-5472.CAN-08-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin-Lluesma S, Schaeffer C, Robert EI, van Breugel PC, Leupin O, Hantz O, Strubin M. Hepatitis B virus X protein affects S phase progression leading to chromosome segregation defects by binding to damaged DNA binding protein 1. Hepatology. 2008;48:1467–1476. doi: 10.1002/hep.22542. [DOI] [PubMed] [Google Scholar]
- 76.Neuveut C, Wei Y, Buendia MA. Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol. 2010;52:594–604. doi: 10.1016/j.jhep.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 77.Rawat S, Clippinger AJ, Bouchard MJ. Modulation of apoptotic signaling by the hepatitis B virus X protein. Viruses. 2012;4:2945–2972. doi: 10.3390/v4112945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clippinger AJ, Gearhart TL, Bouchard MJ. Hepatitis B virus X protein modulates apoptosis in primary rat hepatocytes by regulating both NF-kappaB and the mitochondrial permeability transition pore. J Virol. 2009;83:4718–4731. doi: 10.1128/JVI.02590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rawat S, Bouchard MJ. The hepatitis B virus (HBV) HBx protein activates AKT to simultaneously regulate HBV replication and hepatocyte survival. J Virol. 2015;89:999–1012. doi: 10.1128/JVI.02440-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Su F, Theodosis CN, Schneider RJ. Role of NF-KB and Myc proteins in apoptosis induced by hepatitis B virus HBx protein. J Virol. 2001;75:215–255. doi: 10.1128/JVI.75.1.215-225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee YI, Hwang JM, Im JH, Lee YI, Kim NS, Kim DG, Yu DY, Moon HB, Park SK. Human hepatitis B virus-X protein alters mitochondrial function and physiology in human liver cells. J Biol Chem. 2004;279:15460–15471. doi: 10.1074/jbc.M309280200. [DOI] [PubMed] [Google Scholar]
- 82.Zhang H, Huang C, Wang Y, Lu Z, Zhuang N, Zhao D, He J, Shi L. Hepatitis B Virus X Protein Sensitizes TRAIL-Induced Hepatocyte Apoptosis by Inhibiting the E3 Ubiquitin Ligase A20. PLoS One. 2015;10:e0127329. doi: 10.1371/journal.pone.0127329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liang X, Liu Y, Zhang Q, Gao L, Han L, Ma C, Zhang L, Chen YH, Sun W. Hepatitis B virus sensitizes hepatocytes to TRAIL-induced apoptosis through Bax. J Immunol. 2007;178:503–510. doi: 10.4049/jimmunol.178.1.503. [DOI] [PubMed] [Google Scholar]
- 84.Miao J, Chen GG, Chun SY, Lai PP. Hepatitis B virus X protein induces apoptosis in hepatoma cells through inhibiting Bcl-xL expression. Cancer Lett. 2006;236:115–124. doi: 10.1016/j.canlet.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 85.Chen J, Siddiqui A. Hepatitis B virus protein stimulates the mitochondrial translocation of Raf-1 via oxidative stress. J VIrol. 2007;81:6757–6760. doi: 10.1128/JVI.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim SJ, Khan M, Quan J, Till A, Subramani S, Siddiqui A. Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PLoS Pathog. 2013;9:e1003722. doi: 10.1371/journal.ppat.1003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Waris G, Huh KW, Siddiqui A. Mitochondrially associated hepatitis B virus X protein consitutively activates transcription factors STAT-3 and NF-κB via oxidative stress. Mol Cell Biol. 2001;21:7721–7730. doi: 10.1128/MCB.21.22.7721-7730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88•.Clippinger AJ, Bouchard MJ. The hepatitis B virus HBx protein localizes to mitochondria in primary rat hepatocytes and modulates mitochondrial membrane potential. J Virol. 2008;82:6798–6811. doi: 10.1128/JVI.00154-08. First demonstration that HBx balances anti- and pro-apoptotic signals in primary hepatocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bar-Yishay I, Shaul Y, Shlomai A. Hepatocyte metabolic signalling pathways and regulation of hepatitis B virus expression. Liver Int. 2011;31:282–290. doi: 10.1111/j.1478-3231.2010.02423.x. [DOI] [PubMed] [Google Scholar]
- 90.Shin HJ, Park YH, Kim SU, Moon HB, Park DS, Han YH, Lee CH, Lee DS, Song IS, Lee DH, Kim M, Kim NS, Kim DG, Kim JM, Kim SK, Kim YN, Kim SS, Choi CS, Kim YB, Yu DY. Hepatitis B virus X protein regulates hepatic glucose homeostasis via activation of inducible nitric oxide synthase. J Biol Chem. 2011;286:29872–29881. doi: 10.1074/jbc.M111.259978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91•.Bagga S, Rawat S, Ajenjo M, Bouchard MJ. Hepatitis B virus (HBV) X protein-mediated regulation of hepatocyte metabolic pathways affects viral replication. Virology. 2016;498:9–22. doi: 10.1016/j.virol.2016.08.006. First demonstration that HBx balances anabolic and catabolic signals to regulate HBV replication in hepatocytes. [DOI] [PubMed] [Google Scholar]
- 92.Golob-Schwarzl N, Krassnig S, Toeglhofer AM, Park YN, Gogg-Kamerer M, Vierlinger K, Schroder F, Rhee H, Schicho R, Fickert P, Haybaeck J. New liver cancer biomarkers: PI3K/AKT/mTOR pathway members and eukaryotic translation initiation factors. Eur J Cancer. 2017;83:56–70. doi: 10.1016/j.ejca.2017.06.003. [DOI] [PubMed] [Google Scholar]