Figure 2. Nuclear envelope rupture and genome instability.
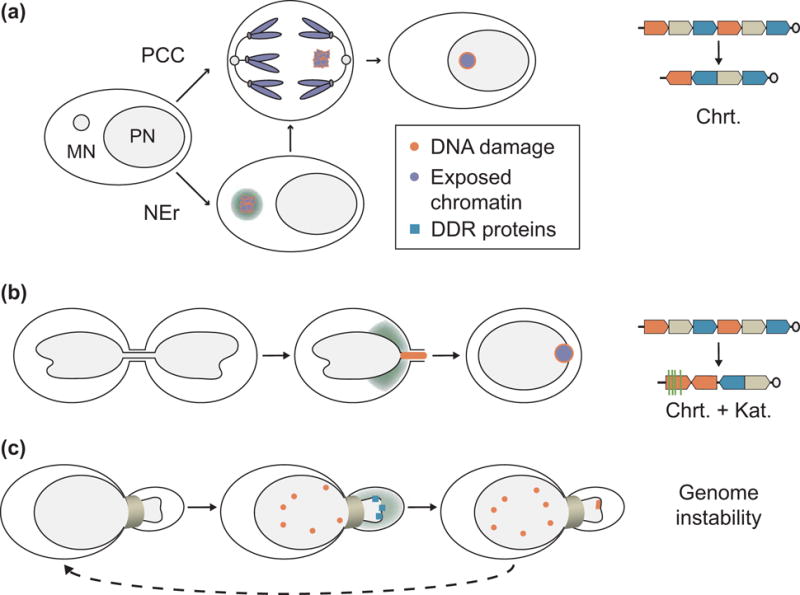
(a) Micronucleus mechanism of chromothripsis. Micronucleation of a chromosome leads to DNA damage and fragmentation after DNA replication initiation, either by NE rupture in interphase, or by premature chromatin condensation in mitosis. Chromothriptic are visible in the next cell cycle, likely as a result of the chromatin being reincorporated in the nucleus and exposed to DNA damage repair proteins. (b) Chromothripsis and kataegis after chromatin bridge resolution. Chromatin bridge breakage is preceded by NE rupture and results in DNA damage and both chromothriptic chromatin rearrangements and APOBEC-associated kataegis. (c) Transient NE rupture in cells migrating through narrow channels is associated with increased DNA damage and mislocalization of DNA damage repair proteins. Repeated migration leads to increased genetic diversity. APOBEC = apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like. Chrt = chromothriptic chromosome rearrangements; DDR = DNA damage repair; Kat = kataegis; NEr = nuclear envelope rupture; PPC = premature chromosome condensation.
