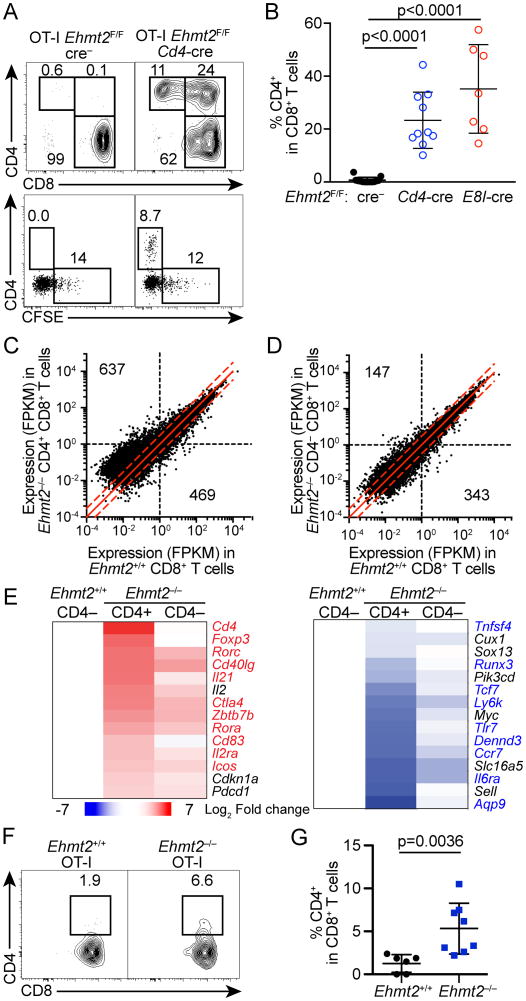
Figure 2. G9a is required to maintain silencing of helper lineage genes in CD8 T cells during lymphopenia- or tumor Ag-driven proliferation.
(A, B) CD4 expression and CFSE dilution of CD8 T cells in PBMC of Tcrb–/–Tcrd–/– mice that received Ehmt2–/– or Ehmt2+/+ OT-I T cells 4 weeks prior to the analysis. Data are pooled from 3 experiments in which one donor of each genotype was transferred into 2-3 recipients. (C, D) RNA-seq analysis of CD4+ CD8+ Ehmt2–/–, CD4– CD8+ Ehmt2–/– and CD4– CD8+ Ehmt2+/+ OT-I T cells harvested from Tcrb–/–Tcrd–/– mice 4 weeks after transfer. Quantification of genes with ≥1 FPKM in Ehmt2–/– or Ehmt2+/+ samples and >2-fold difference in expression is indicated for each genotype. Dashed red lines: 2-fold change between genotypes. (E) Heat maps showing genes differentially expressed between CD4+ CD8+ or CD4– CD8+ Ehmt2–/– and control Ehmt2+/+ CD8 T cells. Values represent the log2 fold change of the mean of 2-4 mice compared to Ehmt2+/+ CD8 T cells. Highlighted genes represent genes differentially expressed between CD4 and CD8 memory T cells from ImmGen datasets. (F, G) Expression of CD4 of OT-I T cells in the lymph node draining transplanted E.G7-OVA tumors. n=6-8 in 2 experiments.