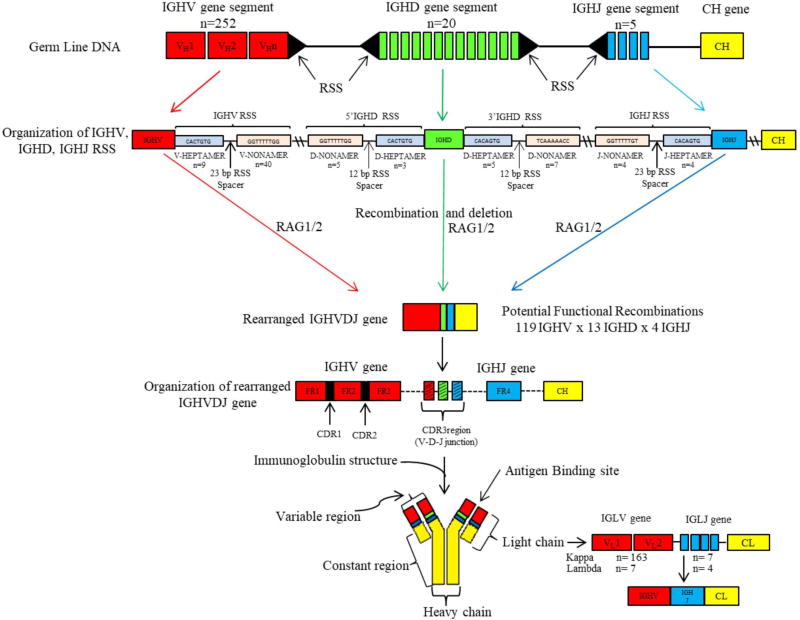
FIG. 2.
Structural organization of IGH and IGL in the rat. The above diagram demonstrates the organization of IGH gene segments position on chromosome 6q32-33 on the IGH locus of the Brown Norway (BN) rat. The IGH locus consists of an IGH variable region and heavy constant (CH) region. IGH variable region is grouped into three germline elements: IGHV, IGHD, and IGHJ gene segments respectively. Also depicted in the above figure are the recognition signal sequenced (RSS) flanking each of IGHV, IGHJ on one side, and IGHD on both sides. RSS consist of conserved heptamer and nonamer sequences, separated from each other by either a 12 or a 23 base-pair (bp) RSS spacer. Recombination of germline IGHV, IGHD, IGHJ (VDJ rearrangement) gene segments follow a 12/23 rule, i.e., VDJ rearrangement occurs only between a 12 bp RSS spacer and a 23 bp RSS spacer. Recombination activating Genes (RAG1/2) bind to RSS’s initiating VDJ rearrangement that results in the sequential random joining of a single gene segment of IGHD to IGHJ and IGHV to IGHDJ forming a rearranged IGHVDJ gene that encodes for the variable part of the immunoglobulin molecule. The IGHV gene is further arranged into CDR1 and CDR2 that are separated from each other by conserved framework regions (FR1, FR2, FR3). FR4 is completely encoded by the IGHJ gene. The CDR3 (junctional) region is the product of VDJ rearrangement. CH chain genes are separately joined by splicing to the rearranged IGHVDJ gene. The total number of gene segments, heptamer and nonamer sequences are designated by “n.”