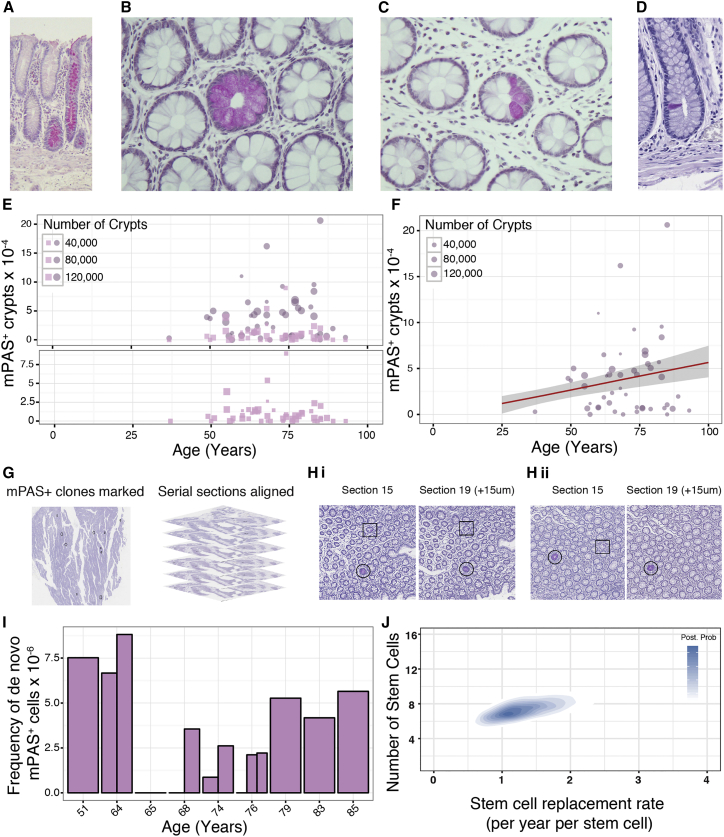
Figure 1.
Identification and Quantification of mPAS+ Clones
(A) Longitudinally sectioned sporadic mPAS+ wholly populated crypts (WPC).
(B and C) Sporadic WPC (B) and partially populated crypts (PPC) (C) within en face tissue sections.
(D) Single mPAS+ cell within a crypt.
(E) Frequencies of mPAS+ WPC (circles) and PPC (squares) plotted against patient age. Bottom panel shows PPC only on expanded y axis.
(F) Regression analysis showing ΔCfix plotted in red at 5.85 × 10−6 per year with 95% ME in gray.
(G) mPAS+ clones are marked within processed images in black before serial sections are aligned to enable tracking of clones.
(H) WPC (circles) and PPC (squares) can be traced through aligned serial sections (i), while de novo mutations occurring in TA cells cannot (ii).
(I) Frequency of de novo mPAS+ cells derived for 9 patients plotted by age. Each bar represents a single sample, up to three samples were analyzed per patient. The overall mutation rate (α) was calculated to be 4.44 × 10−6 mutations per mitoses (±2.69 × 106; ME 95%).
(J) Heatmap representing posterior probabilities for the indicated combination of functional stem cell number for crypt (N, y axis) and the rate of stem cell replacement per year per stem cell (λ, x axis). Colors represent posterior probability, white indicating a very low probability that this value underlies the actual dynamics observed, blue indicating a high likelihood. Inference of N and λ in human colonic crypts indicates between 5 and 10 (95% CI; mean = 7) functional stem cells replacing each other at a rate of between 0.65 and 2.7 stem cell replacements per crypt per year (95% CI; mean = 1.3).
See also Figures S1 and S2.