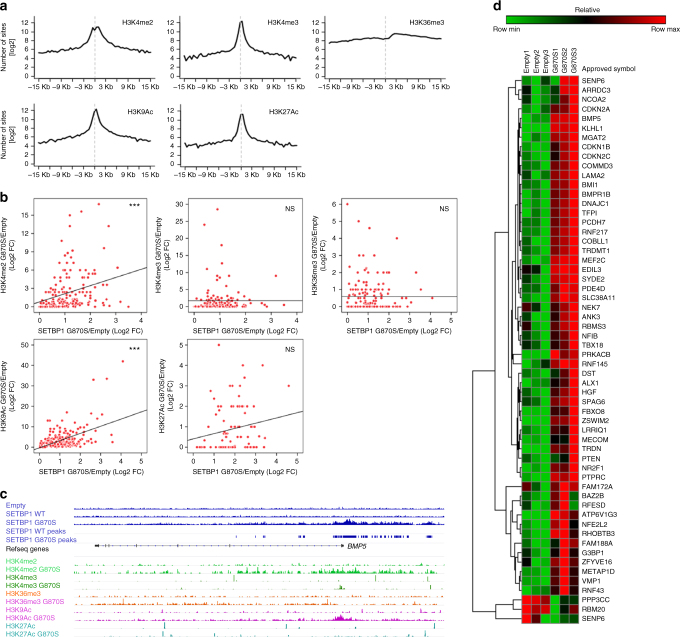
Fig. 3.
SETBP1-mediated epigenetic modulation. a Histone modification ChIP-Seq peak distribution densities plotted according to their distance from gene transcription start sites. b Epigenetic changes resulting from the presence of SETBP1-G870S expressed as function of SETBP1 differential DNA binding (ChIP-Seq G870S/Empty fold change in x axis) vs. histone modification differential enrichment (G870S/Empty fold change in y axis). ***p < 0.001 c SETBP1 ChIP-Seq coverage track and peak alignment to the hg19 reference genome are superimposed to the different histone methylation ChIP-Seq coverage tracks within the BMP5 locus. d Gene expression heatmap of the subset of SETBP1 targets harboring increased H3K4me2 and H3K9ac activation marks generated on three Empty and three SETBP1-G870S clones