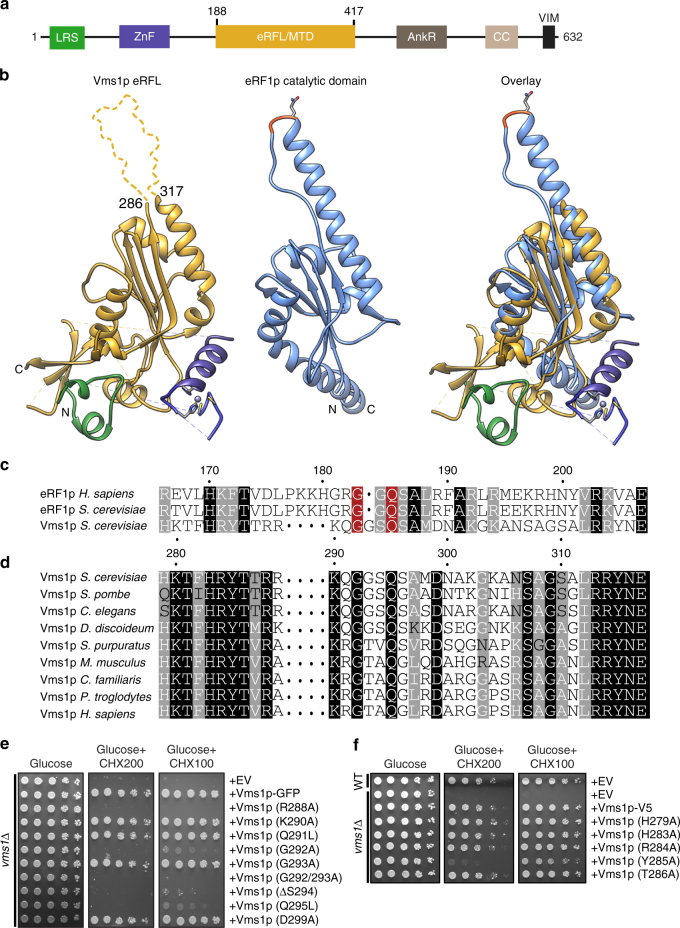
Fig. 3.
Vms1p is structurally homologous to tRNA hydrolases. a Domain structure of Vms1p. LRS leucine-rich sequence, ZnF zinc finger, MTD/eRFL mitochondrial targeting domain/eRF1-like, AnkR ankryin repeat, CC coil–coil, VIM VCP-interacting motif. Residues 188–417 represent the MTD/eRFL boundaries. b Structural alignment of Vms1p (left, 5WHG31) and eRF1p (middle, 3JAHii35, residues 144–280). Dashed lines indicate connections made by residues that are not resolved in the Vms1p crystal structure. The GGQ (red) loop of eRF1p is ordered in the ribosome-bound structure shown here. c Sequence alignment of Vms1p and eRF1p. White letters with gray, black, or red background indicates similarity, identity, or GxxQ residues, respectively. d Sequence alignment of Vms1p orthologs across the GxxQ region. Coloring as in c. e, f Serial dilutions of indicated strains were spotted on media containing glucose or glucose supplemented with cycloheximide (CHX). EV empty vector