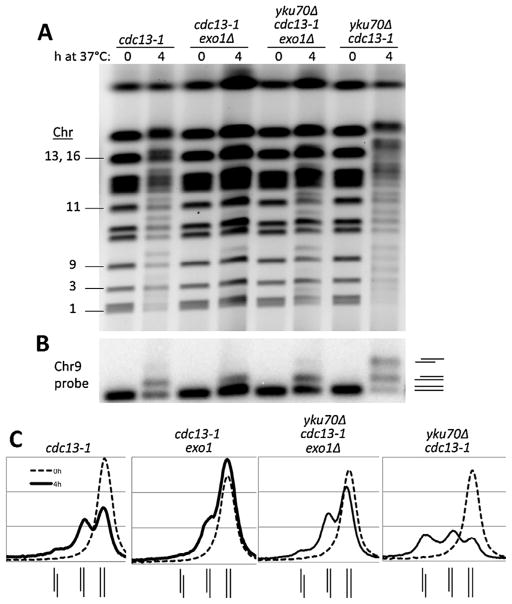
Fig. 2.
The role of Exo1 in global resection at telomeres of cdc13-1 and yku70Δ cdc13-1 mutants. (A) Cells with indicated genotypes in the CG379 strain background grown to late log at 20 °C (YPDA+SRB) were diluted to fresh medium and shifted to 37 °C. Chromosomal DNA was examined using the PFGE protocols (see Material and Methods). (B) Southern of the gel shown in panel (A) using a 32P labeled Chr 9 probe. (C) The signal intensity profiles of 0 h and 4 h timepoints from the Southern from panel (B) indicate the distribution of 0-, 1-, and 2-end resected forms of Chr 9 for each strain. The line diagrams beneath each graph indicate the positions (left to right) of the 2-end, 1-end, and 0-end resection peaks.