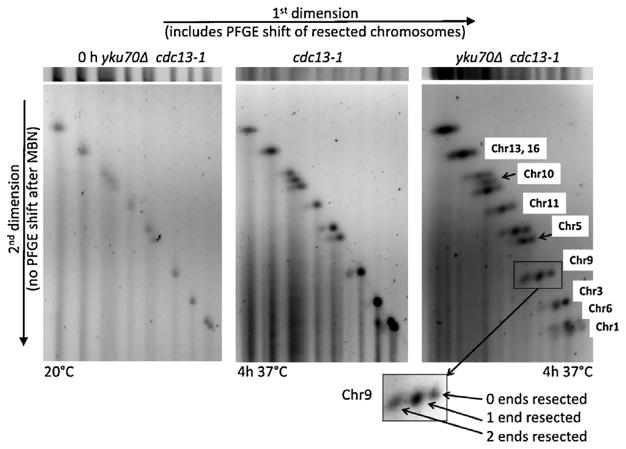
Fig. 3.
2-D PFGE analysis along with nuclease treatment identifies gobal 0-, 1-, and 2-end resection along with chromosomal differences. Lanes (horizontal slices) in the first dimension were cut out from the PFGE gel and exposed to mung bean nuclease to remove ssDNA tails. The lane is then subjected to PFGE in the second dimension. The bands of chromosomes with ssDNA tails that were PFGE-shifted in the first dimension do not undergo PFGE-shift in the second dimension. Because of the nuclease digestion, they are shortened compared to the chromosome with no resection and will have greater mobility. Because the 1-end and 2-end resected chromosomes were separated from the 0-end chromosomes, the relative amounts of resected chromosomes can be approximately quantitated, as indicated in the inset.