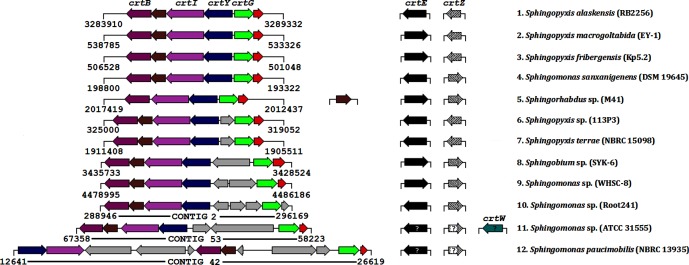
Fig. 1.
Comparison of crt ORFs/loci in 12 strains of Sphingomonadales. Serial numbers of the bacteria are the same as in Table 1. The crt ORF that each colour-coded or shaded arrow represents is indicated on the top. The brown and red arrows represent ORFs encoding the putative LOG and DUF2141 proteins, respectively. The grey arrows represent the separating ORFs that are unlikely to be involved in carotenoid biosynthesis. Numbers at the beginning and end of gene clusters indicate the coordinates within each genome or contig. In the case of crtE and crtZ, arrows pointing to the right indicate that they were located on the plus strand, while those pointing to the left indicate that they were located on the minus strand. The uncertainty of the location of these ORFs on the plus or minus strands in the draft genomes of strains ATCC 31555 and NBRC 13935 is denoted by a ‘?’ within the respective arrows.